Wir bei arbeiten viel mit R und verwenden oft die gleichen kleinen Hilfsfunktionen in unseren Projekten. Diese Funktionen erleichtern unseren Arbeitsalltag, indem sie sich-wiederholende Codeteile reduzieren oder Übersichten über unsere Projekte erstellen.
Um diese Funktionen innerhalb unserer Teams und auch mit anderen zu teilen, habe ich angefangen, sie zu sammeln und habe dann daraus ein R-Paket namens erstellt. Neben der gemeinsamen Nutzung wollte ich auch einige Anwendungsfälle haben, um meine Fähigkeiten zur Fehlersuche und Optimierung zu verbessern. Mit der Zeit wuchs das Paket und es kamen immer mehr Funktionen zusammen. Beim letzten Mal habe ich jede Funktion als Teil eines Adventskalenders vorgestellt. Zum Start unserer neuen Website habe ich alle Funktionen in diesem Kalender zusammengefasst und werde jede aktuelle Funktion aus dem Paket vorstellen.
Die meisten Funktionen wurden entwickelt, als es ein Problem gab und man eine einfache Lösung dafür brauchte. Zum Beispiel war der angezeigte Text zu lang und musste gekürzt werden (siehe evenstrings). Andere Funktionen existieren nur, um sich-wiederholende Aufgaben zu reduzieren – wie das Einlesen mehrerer Dateien des selben Typs (siehe read_files). Daher könnten diese Funktionen auch für Euch nützlich sein!
Um alle Funktionen im Detail zu erkunden, könnt Ihr unser besuchen. Wenn Ihr irgendwelche Vorschläge habt, schickt mir bitte eine E-Mail oder öffnet ein Issue auf GitHub!
1. char_replace
Dieser kleine Helfer ersetzt Sonderzeichen (wie z. B. den Umlaut „ä“) durch ihre Standardentsprechung (in diesem Fall „ae“). Es ist auch möglich, alle Zeichen in Kleinbuchstaben umzuwandeln, Leerzeichen zu entfernen oder Leerzeichen und Bindestriche durch Unterstriche zu ersetzen.
Schauen wir uns ein kleines Beispiel mit verschiedenen Settings an:
x <- " Élizàldë-González Strasse"
char_replace(x, to_lower = TRUE)[1] "elizalde-gonzalez strasse"char_replace(x, to_lower = TRUE, to_underscore = TRUE)[1] "elizalde_gonzalez_strasse"char_replace(x, to_lower = FALSE, rm_space = TRUE, rm_dash = TRUE)[1] "ElizaldeGonzalezStrasse"2. checkdir
Dieser kleine Helfer prüft einen gegebenen Ordnerpfad auf Existenz und erstellt ihn bei Bedarf.
checkdir(path = "testfolder/subfolder")Intern gibt es nur eine einfache if-Anweisung, die die R-Basisfunktionen file.exists() und dir.create(). kombiniert.
3. clean_gc
Dieser kleine Helfer gibt den Speicher von unbenutzten Objekten frei. Nun, im Grunde ruft es einfach gc() ein paar Mal auf. Ich habe das vor einiger Zeit für ein Projekt benutzt, bei dem ich mit riesigen Datendateien gearbeitet habe. Obwohl wir das Glück hatten, einen großen Server mit 500 GB RAM zu haben, stießen wir bald an seine Grenzen. Da wir in der Regel mehrere Prozesse parallelisieren, mussten wir jedes Bit und jedes Byte des Arbeitsspeichers nutzen, das wir bekommen konnten. Anstatt also viele Zeilen wie diese zu haben:
gc();gc();gc();gc()… habe ich clean_gc() der Einfachheit halber geschrieben. Intern wird gc() so lange aufgerufen, wie es Speicher gibt, der freigegeben werden muss.
Some further thoughts
Es gibt einige Diskussionen über den Garbage Collector gc() und seine Nützlichkeit. Wenn Ihr mehr darüber erfahren wollt, schlage ich vor, dass Ihr Euch die anseht. Ich weiß, dass R selbst bei Bedarf Speicher freigibt, aber ich bin mir nicht sicher, was passiert, wenn Ihr mehrere R-Prozesse habt. Können sie den Speicher von anderen Prozessen leeren? Wenn Ihr dazu etwas mehr wisst, lasst es mich wissen!
4. count_na
Dieser kleine Helfer zählt fehlende Werte innerhalb eines Vektors.
x <- c(NA, NA, 1, NaN, 0)
count_na(x)3Intern gibt es nur ein einfaches sum(is.na(x)), das die NA-Werte zählt. Wenn Ihr den Mittelwert statt der Summe wollt, könnt Ihr prop = TRUE setzen.
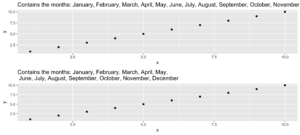
5. evenstrings
Dieser kleine Helfer zerlegt eine gegebene Zeichenkette in kleinere Teile mit einer festen Länge. Aber warum? Nun, ich brauchte diese Funktion beim Erstellen eines Plots mit einem langen Titel. Der Text war zu lang für eine Zeile und anstatt ihn einfach abzuschneiden oder über die Ränder laufen zu lassen, wollte ich ihn schön trennen.
Bei einer langen Zeichenkette wie…
long_title <- c("Contains the months: January, February, March, April, May, June, July, August, September, October, November, December")…wollen wir sie nach split = "," mit einer maximalen Länge von char = 60 aufteilen.
short_title <- evenstrings(long_title, split = ",", char = 60)Die Funktion hat zwei mögliche Ausgabeformate, die durch Setzen von newlines = TRUE oder FALSE gewählt werden können:
- eine Zeichenkette mit Zeilentrennzeichen
\n - ein Vektor mit jedem Unterteil.
Ein anderer Anwendungsfall könnte eine Nachricht sein, die mit cat() auf der Konsole ausgegeben wird:
cat(long_title)Contains the months: January, February, March, April, May, June, July, August, September, October, November, Decembercat(short_title)Contains the months: January, February, March, April, May,
June, July, August, September, October, November, DecemberCode for plot example
p1 <- ggplot(data.frame(x = 1:10, y = 1:10),
aes(x = x, y = y)) +
geom_point() +
ggtitle(long_title)
p2 <- ggplot(data.frame(x = 1:10, y = 1:10),
aes(x = x, y = y)) +
geom_point() +
ggtitle(short_title)
multiplot(p1, p2)6. get_files
Dieser kleine Helfer macht das Gleiche wie die „Find in files „ Suche in RStudio. Sie gibt einen Vektor mit allen Dateien in einem bestimmten Ordner zurück, die das Suchmuster enthalten. In Eurem täglichen Arbeitsablauf würdet Ihr normalerweise die Tastenkombination SHIFT+CTRL+F verwenden. Mit get_files() könnt Ihr diese Funktionen in Euren Skripten nutzen.
7. get_network
Das Ziel dieses kleinen Helfers ist es, die Verbindungen zwischen R-Funktionen innerhalb eines Projekts als Flussdiagramm zu visualisieren. Dazu ist die Eingabe ein Verzeichnispfad zur Funktion oder eine Liste mit den Funktionen und die Ausgaben sind eine Adjazenzmatrix und ein Graph-Objekt. Als Beispiel verwenden wir diesen Ordner mit einigen Spielzeugfunktionen:
net <- get_network(dir = "flowchart/R_network_functions/", simplify = FALSE)
g1 <- net$igraphInput
Es gibt fünf Parameter, um mit der Funktion zu interagieren:
- ein Pfad
dir, der durchsucht werden soll. - ein Zeichenvektor
Variationenmit der Definitionszeichenfolge der Funktion – die Vorgabe istc(" <- function", "<- function", "<-function"). - ein „Muster“, eine Zeichenkette mit dem Dateisuffix – die Vorgabe ist
"\\.R$". - ein boolesches
simplify, das Funktionen ohne Verbindungen aus der Darstellung entfernt. - eine benannte Liste
all_scripts, die eine Alternative zudirist. Diese Liste wird hauptsächlich nur zu Testzwecken verwendet.
Für eine normale Verwendung sollte es ausreichen, einen Pfad zum Projektordner anzugeben.
Output
Der gegebene Plot zeigt die Verbindungen der einzelnen Funktionen (Pfeile) und auch die relative Größe des Funktionscodes (Größe der Punkte). Wie bereits erwähnt, besteht die Ausgabe aus einer Adjazenzmatrix und einem Graph-Objekt. Die Matrix enthält die Anzahl der Aufrufe für jede Funktion. Das Graph-Objekt hat die folgenden Eigenschaften:
- Die Namen der Funktionen werden als Label verwendet.
- Die Anzahl der Zeilen jeder Funktion (ohne Kommentare und Leerzeilen) wird als Größe gespeichert.
- Der Ordnername des ersten Ordners im Verzeichnis.
- Eine Farbe, die dem Ordner entspricht.
Mit diesen Eigenschaften können Sie die Netzwerkdarstellung zum Beispiel wie folgt verbessern:
library(igraph)
# create plots ------------------------------------------------------------
l <- layout_with_fr(g1)
colrs <- rainbow(length(unique(V(g1)$color)))
plot(g1,
edge.arrow.size = .1,
edge.width = 5*E(g1)$weight/max(E(g1)$weight),
vertex.shape = "none",
vertex.label.color = colrs[V(g1)$color],
vertex.label.color = "black",
vertex.size = 20,
vertex.color = colrs[V(g1)$color],
edge.color = "steelblue1",
layout = l)
legend(x = 0,
unique(V(g1)$folder), pch = 21,
pt.bg = colrs[unique(V(g1)$color)],
pt.cex = 2, cex = .8, bty = "n", ncol = 1)
Dieser kleine Helfer gibt Indizes von wiederkehrenden Mustern zurück. Es funktioniert sowohl mit Zahlen als auch mit Zeichen. Alles, was es braucht, ist ein Vektor mit den Daten, ein Muster, nach dem gesucht werden soll, und eine Mindestanzahl von Vorkommen.
Lasst uns mit dem folgenden Code einige Zeitreihendaten erstellen.
library(data.table)
# random seed
set.seed(20181221)
# number of observations
n <- 100
# simulationg the data
ts_data <- data.table(DAY = 1:n, CHANGE = sample(c(-1, 0, 1), n, replace = TRUE))
ts_data[, VALUE := cumsum(CHANGE)]Dies ist nichts anderes als ein Random Walk, da wir zwischen dem Abstieg (-1), dem Anstieg (1) und dem Verbleib auf demselben Niveau (0) wählen. Unsere Zeitreihendaten sehen folgendermaßen aus:
Angenommen, wir wollen die Datumsbereiche wissen, in denen es an mindestens vier aufeinanderfolgenden Tagen keine Veränderung gab.
ts_data[, get_sequence(x = CHANGE, pattern = 0, minsize = 4)] min max
[1,] 45 48
[2,] 65 69Wir können auch die Frage beantworten, ob sich das Muster „down-up-down-up“ irgendwo wiederholt:
ts_data[, get_sequence(x = CHANGE, pattern = c(-1,1), minsize = 2)] min max
[1,] 88 91Mit diesen beiden Eingaben können wir unseren Plot ein wenig aktualisieren, indem wir etwas geom_rect hinzufügen!
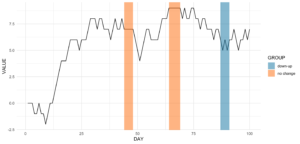
Code for the plot
rect <- data.table(
rbind(ts_data[, get_sequence(x = CHANGE, pattern = c(0), minsize = 4)],
ts_data[, get_sequence(x = CHANGE, pattern = c(-1,1), minsize = 2)]),
GROUP = c("no change","no change","down-up"))
ggplot(ts_data, aes(x = DAY, y = VALUE)) +
geom_line() +
geom_rect(data = rect,
inherit.aes = FALSE,
aes(xmin = min - 1,
xmax = max,
ymin = -Inf,
ymax = Inf,
group = GROUP,
fill = GROUP),
color = "transparent",
alpha = 0.5) +
scale_fill_manual(values = statworx_palette(number = 2, basecolors = c(2,5))) +
theme_minimal()9. intersect2
Dieser kleine Helfer gibt den Schnittpunkt mehrerer Vektoren oder Listen zurück. Ich habe diese Funktion gefunden, fand sie recht nützlich und habe sie ein wenig angepasst.
intersect2(list(c(1:3), c(1:4)), list(c(1:2),c(1:3)), c(1:2))[1] 1 2Intern wird das Problem, die Schnittmenge zu finden, rekursiv gelöst, wenn ein Element eine Liste ist, und dann schrittweise mit dem nächsten Element.
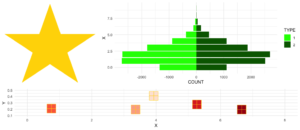
10. multiplot
Dieses kleine Hilfsmittel kombiniert mehrere ggplots zu einem Plot. Dies ist eine Funktion aus .
Ein Vorteil gegenüber facets ist, dass man nicht alle Daten für alle Plots in einem Objekt benötigt. Auch kann man jeden einzelnen Plot frei erstellen – was manchmal auch ein Nachteil sein kann.
Mit dem Parameter layout könnt Ihr mehrere Plots mit unterschiedlichen Größen anordnen. Nehmen wir an, Ihr habt drei Plots und wollt sie wie folgt anordnen:
1 2 2
1 2 2
3 3 3Bei multiplot läuft es auf Folgendes hinaus:
multiplot(plotlist = list(p1, p2, p3),
layout = matrix(c(1,2,2,1,2,2,3,3,3), nrow = 3, byrow = TRUE))Code for plot example
# star coordinates
c1 = cos((2*pi)/5)
c2 = cos(pi/5)
s1 = sin((2*pi)/5)
s2 = sin((4*pi)/5)
data_star <- data.table(X = c(0, -s2, s1, -s1, s2),
Y = c(1, -c2, c1, c1, -c2))
p1 <- ggplot(data_star, aes(x = X, y = Y)) +
geom_polygon(fill = "gold") +
theme_void()
# tree
set.seed(24122018)
n <- 10000
lambda <- 2
data_tree <- data.table(X = c(rpois(n, lambda), rpois(n, 1.1*lambda)),
TYPE = rep(c("1", "2"), each = n))
data_tree <- data_tree[, list(COUNT = .N), by = c("TYPE", "X")]
data_tree[TYPE == "1", COUNT := -COUNT]
p2 <- ggplot(data_tree, aes(x = X, y = COUNT, fill = TYPE)) +
geom_bar(stat = "identity") +
scale_fill_manual(values = c("green", "darkgreen")) +
coord_flip() +
theme_minimal()
# gifts
data_gifts <- data.table(X = runif(5, min = 0, max = 10),
Y = runif(5, max = 0.5),
Z = sample(letters[1:5], 5, replace = FALSE))
p3 <- ggplot(data_gifts, aes(x = X, y = Y)) +
geom_point(aes(color = Z), pch = 15, size = 10) +
scale_color_brewer(palette = "Reds") +
geom_point(pch = 12, size = 10, color = "gold") +
xlim(0,8) +
ylim(0.1,0.5) +
theme_minimal() +
theme(legend.position="none")
11. na_omitlist
Dieser kleine Helfer entfernt fehlende Werte aus einer Liste.
y <- list(NA, c(1, NA), list(c(5:6, NA), NA, "A"))Es gibt zwei Möglichkeiten, die fehlenden Werte zu entfernen, entweder nur auf der ersten Ebene der Liste oder innerhalb jeder Unterebene.
na_omitlist(y, recursive = FALSE)[[1]]
[1] 1 NA
[[2]]
[[2]][[1]]
[1] 5 6 NA
[[2]][[2]]
[1] NA
[[2]][[3]]
[1] "A"na_omitlist(y, recursive = TRUE)[[1]]
[1] 1
[[2]]
[[2]][[1]]
[1] 5 6
[[2]][[2]]
[1] "A"12. %nin%
Dieser kleine Helfer ist eine reine Komfortfunktion. Sie ist einfach dasselbe wie der negierte %in%-Operator, wie Ihr unten sehen könnt. Aber meiner Meinung nach erhöht sie die Lesbarkeit des Codes.
all.equal( c(1,2,3,4) %nin% c(1,2,5),
!c(1,2,3,4) %in% c(1,2,5))[1] TRUEDieser Operator hat es auch in einige andere Pakete geschafft – .
13. object_size_in_env
Dieser kleine Helfer zeigt eine Tabelle mit der Größe jedes Objekts in der vorgegebenen Umgebung an.
Wenn Ihr in einer Situation seid, in der Ihr viel gecodet habt und Eure Umgebung nun ziemlich unübersichtlich ist, hilft Euch object_size_in_env, die großen Fische in Bezug auf den Speicherverbrauch zu finden. Ich selbst bin ein paar Mal auf dieses Problem gestoßen, als ich mehrere Ausführungen meiner Modelle in einem Loop durchlaufen habe. Irgendwann wurden die Sitzungen ziemlich groß im Speicher und ich wusste nicht, warum! Mit Hilfe von object_size_in_env und etwas Degubbing konnte ich das Objekt ausfindig machen, das dieses Problem verursachte, und meinen Code entsprechend anpassen.
Zuerst wollen wir eine Umgebung mit einigen Variablen erstellen.
# building an environment
this_env <- new.env()
assign("Var1", 3, envir = this_env)
assign("Var2", 1:1000, envir = this_env)
assign("Var3", rep("test", 1000), envir = this_env)Um die Größeninformationen unserer Objekte zu erhalten, wird intern format(object.size()) verwendet. Mit der Einheit kann das Ausgabeformat geändert werden (z.B. "B", "MB" oder "GB").
# checking the size
object_size_in_env(env = this_env, unit = "B") OBJECT SIZE UNIT
1: Var3 8104 B
2: Var2 4048 B
3: Var1 56 B14. print_fs
Dieser kleine Helfer gibt die Ordnerstruktur eines gegebenen Pfades zurück. Damit kann man z.B. eine schöne Übersicht in die Dokumentation eines Projektes oder in ein Git einbauen. Im Sinne der Automatisierung könnte diese Funktion nach einer größeren Änderung Teile in einer Log- oder News-Datei ändern.
Wenn wir uns das gleiche Beispiel anschauen, das wir für die Funktion get_network verwendet haben, erhalten wir folgendes:
print_fs("~/flowchart/", depth = 4)1 flowchart
2 ¦--create_network.R
3 ¦--getnetwork.R
4 ¦--plots
5 ¦ ¦--example-network-helfRlein.png
6 ¦ °--improved-network.png
7 ¦--R_network_functions
8 ¦ ¦--dataprep
9 ¦ ¦ °--foo_01.R
10 ¦ ¦--method
11 ¦ ¦ °--foo_02.R
12 ¦ ¦--script_01.R
13 ¦ °--script_02.R
14 °--README.md Mit depth können wir einstellen, wie tief wir unsere Ordner durchforsten wollen.
15. read_files
Dieser kleine Helfer liest mehrere Dateien des selben Typs ein und fasst sie zu einer data.table zusammen. Welche Art von Dateilesefunktion verwendet werden soll, kann mit dem Argument FUN ausgewählt werden.
Wenn Sie eine Liste von Dateien haben, die alle mit der gleichen Funktion eingelesen werden sollen (z.B. read.csv), können Sie statt lapply und rbindlist nun dies verwenden:
read_files(files, FUN = readRDS)
read_files(files, FUN = readLines)
read_files(files, FUN = read.csv, sep = ";")Intern verwendet es nur lapply und rbindlist, aber man muss es nicht ständig eingeben. Die read_files kombiniert die einzelnen Dateien nach ihren Spaltennamen und gibt eine data.table zurück. Warum data.table? Weil ich es mag. Aber lassen Sie uns nicht das Fass von data.table vs. dplyr aufmachen (zum Fass…).
16. save_rds_archive
Dieser kleine Helfer ist ein Wrapper um die Basis-R-Funktion saveRDS() und prüft, ob die Datei, die Ihr zu speichern versucht, bereits existiert. Wenn ja, wird die bestehende Datei umbenannt / archiviert (mit einem Zeitstempel), und die „aktualisierte“ Datei wird unter dem angegebenen Namen gespeichert. Das bedeutet, dass vorhandener Code, der davon abhängt, dass der Dateiname konstant bleibt (z.B. readRDS()-Aufrufe in anderen Skripten), weiterhin funktionieren wird, während eine archivierte Kopie der – ansonsten überschriebenen – Datei erhalten bleibt.
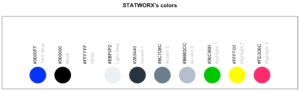
17. sci_palette
Dieser kleine Helfer liefert eine Reihe von Farben, die wir bei statworx häufig verwenden. Wenn Ihr Euch also – so wie ich – nicht an jeden Hex-Farbcode erinnern könnt, den Ihr braucht, könnte das helfen. Natürlich sind das unsere Farben, aber Ihr könnt es auch mit Eurer eigenen Farbpalette umschreiben. Aber der Hauptvorteil ist die Plot-Methode – so könnt Ihr die Farbe sehen, anstatt nur den Hex-Code zu lesen.
So seht Ihr, welcher Hexadezimalcode welcher Farbe entspricht und wofür Ihr ihn verwenden könnt.
sci_palette(scheme = "new")Tech Blue Black White Light Grey Accent 1 Accent 2 Accent 3
"#0000FF" "#000000" "#FFFFFF" "#EBF0F2" "#283440" "#6C7D8C" "#B6BDCC"
Highlight 1 Highlight 2 Highlight 3
"#00C800" "#FFFF00" "#FE0D6C"
attr(,"class")
[1] "sci"Wie bereits erwähnt, gibt es eine Methode plot(), die das folgende Bild ergibt.
plot(sci_palette(scheme = "new"))
18. statusbar
Dieser kleine Helfer gibt einen Fortschrittsbalken in der Konsole für Schleifen aus.
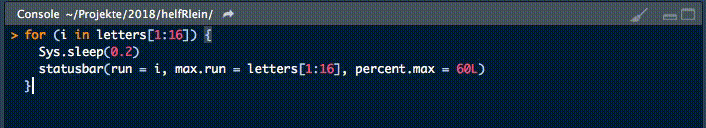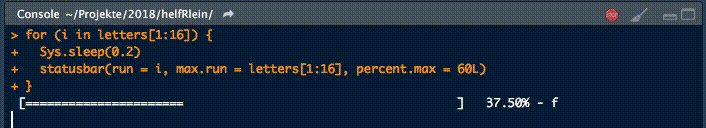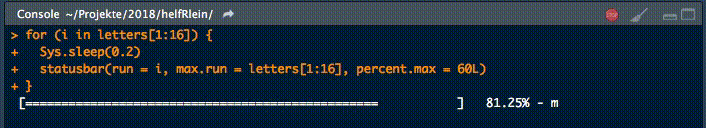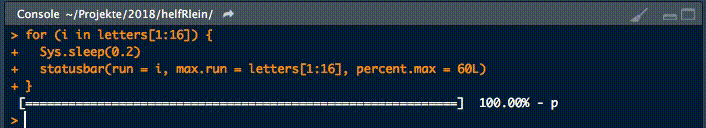
Es gibt zwei notwendige Parameter, um diese Funktion zu füttern:
runist entweder der Iterator oder seine Nummermax.runist entweder alle möglichen Iteratoren in der Reihenfolge, in der sie verarbeitet werden, oder die maximale Anzahl von Iterationen.
So könnte es zum Beispiel run = 3 und max.run = 16 oder run = "a" und max.run = Buchstaben[1:16] sein.
Außerdem gibt es zwei optionale Parameter:
percent.maxbeeinflusst die Breite des Fortschrittsbalkensinfoist ein zusätzliches Zeichen, das am Ende der Zeile ausgegeben wird. Standardmäßig ist esrun.
Ein kleiner Nachteil dieser Funktion ist, dass sie nicht mit parallelen Prozessen arbeitet. Wenn Ihr einen Fortschrittsbalken haben wollt, wenn Ihr apply Funktionen benutzt, schaut Euch an.
19. statworx_palette
Dieses kleine Hilfsmittel ist eine Ergänzung zu sci_palette(). Wir haben die Farben 1, 2, 3, 5 und 10 ausgewählt, um eine flexible Farbpalette zu erstellen. Wenn Sie 100 verschiedene Farben benötigen – sagen Sie nichts mehr!
Im Gegensatz zu sci_palette() ist der Rückgabewert ein Zeichenvektor. Zum Beispiel, wenn Sie 16 Farben wollen:
statworx_palette(16, scheme = "old")[1] "#013848" "#004C63" "#00617E" "#00759A" "#0087AB" "#008F9C" "#00978E" "#009F7F"
[9] "#219E68" "#659448" "#A98B28" "#ED8208" "#F36F0F" "#E45A23" "#D54437" "#C62F4B"Wenn wir nun diese Farben aufzeichnen, erhalten wir einen schönen regenbogenartigen Farbverlauf.
library(ggplot2)
ggplot(plot_data, aes(x = X, y = Y)) +
geom_point(pch = 16, size = 15, color = statworx_palette(16, scheme = "old")) +
theme_minimal()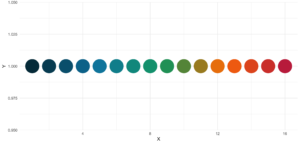
Eine zusätzliche Funktion ist der Parameter reorder, der die Reihenfolge der Farben abtastet, so dass Nachbarn vielleicht etwas besser unterscheidbar sind. Auch wenn Sie die verwendeten Farben ändern wollen, können Sie dies mit basecolors tun.
ggplot(plot_data, aes(x = X, y = Y)) +
geom_point(pch = 16, size = 15,
color = statworx_palette(16, basecolors = c(4,8,10), scheme = "new")) +
theme_minimal()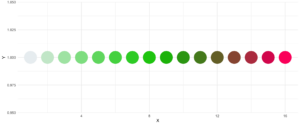
20. strsplit
Dieses kleine Hilfsmittel erweitert die R-Basisfunktion strsplit – daher der gleiche Name! Es ist nun möglich, before, after oder between ein bestimmtes Begrenzungszeichen zu trennen. Im Falle von between müsst ihr zwei Delimiter angeben.
Eine frühere Version dieser Funktion findet Ihr in diesem Blogbeitrag, wo ich die verwendeten regulären Ausdrücke beschreibe, falls Ihr daran interessiert seid.
Hier ist ein kleines Beispiel, wie man das neue strsplit benutzt.
text <- c("This sentence should be split between should and be.")
strsplit(x = text, split = " ")
strsplit(x = text, split = c("should", " be"), type = "between")
strsplit(x = text, split = "be", type = "before")[[1]]
[1] "This" "sentence" "should" "be" "split" "between" "should" "and"
[9] "be."
[[1]]
[1] "This sentence should" " be split between should and be."
[[1]]
[1] "This sentence should " "be split " "between should and "
[4] "be."21. to_na
Dieser kleine Helfer ist nur eine Komfortfunktion. Bei der Datenaufbereitung kann es vorkommen, dass Ihr einen Vektor mit unendlichen Werten wie Inf oder -Inf oder sogar NaN-Werten habt. Solche Werte können (müssen aber nicht!) Eure Auswertungen und Modelle durcheinanderbringen. Aber die meisten Funktionen haben die Tendenz, fehlende Werte zu behandeln. Daher entfernt diese kleine Hilfe solche Werte und ersetzt sie durch NA.
Ein kleines Beispiel, um Euch die Idee zu vermitteln:
test <- list(a = c("a", "b", NA),
b = c(NaN, 1,2, -Inf),
c = c(TRUE, FALSE, NaN, Inf))
lapply(test, to_na)$a
[1] "a" "b" NA
$b
[1] NA 1 2 NA
$c
[1] TRUE FALSE NAEin kleiner Tipp am Rande! Da es je nach den anderen Werten innerhalb eines Vektors verschiedene Arten von NA gibt, solltet Ihr das Format überprüfen, wenn Ihr to_na auf Gruppen oder Teilmengen anwendet.
test <- list(NA, c(NA, "a"), c(NA, 2.3), c(NA, 1L))
str(test)List of 4
$ : logi NA
$ : chr [1:2] NA "a"
$ : num [1:2] NA 2.3
$ : int [1:2] NA 122. trim
Dieser kleine Helfer entfernt führende und nachfolgende Leerzeichen aus einer Zeichenkette. Mit wurde trimws eingeführt, das genau das Gleiche tut. Das zeigt nur, dass es keine schlechte Idee war, eine solche Funktion zu schreiben. 😉
x <- c(" Hello world!", " Hello world! ", "Hello world! ")
trim(x, lead = TRUE, trail = TRUE)[1] "Hello world!" "Hello world!" "Hello world!"Die Parameter lead und trail geben an, ob nur die führenden, die nachfolgenden oder beide Leerzeichen entfernt werden sollen.
Fazit
Ich hoffe, dass euch das helfRlein Package genauso die Arbeit erleichtert, wie uns hier bei statworx. Schreibt uns bei Fragen oder Input zum Package gerne eine Mail an: blog@statworx.com
Im Bereich Data Science – wie der Name schon sagt – ist das Thema Daten, vom Data Cleaning bis hin zum Feature Engineering, einer der Grundpfeiler. Daten zu haben und auszuwerten ist die eine Seite, doch wie kommt man eigentlich an Daten für neue Problemstellungen?
Wenn man Glück hat, werden die Daten, die man benötigt, bereits zur Verfügung gestellt. Sei es über den Download eines ganzen Datensatzes oder die Verwendung einer API. Häufig muss man allerdings auch Informationen von Webseiten selbst zusammentragen – das nennt man Web Scraping. Je nachdem wie oft man Daten scrapen will, ist es von Vorteil, diesen Schritt zu automatisieren.
In diesem Beitrag soll es genau um diese Automatisierung gehen. Ich werde mittels Web Scraping und GitHub Actions an einem Beispiel aufzeigen, wie man sich selbst Datensätze über einen längeren Zeitraum erstellen kann. Dabei soll der Fokus auf den Erfahrungen liegen, die ich in den letzten Monaten gesammelt habe.
Der verwendete Code sowie die bisher gesammelten Daten befinden sich in diesem GitHub Repo.
Suche nach Daten – Ausgangslage
Bei meiner Recherche für den Blogbeitrag über die Benzinpreise, bin ich auch über Daten zur Auslastung der Parkhäuser in Frankfurt am Main gestoßen. Die Beschaffung dieser Daten legte den Grundstein für diesen Beitrag. Nach einigen Überlegungen und zusätzlicher Recherche kamen mir noch weitere thematisch passende Datenquellen in den Sinn:
- Auslastung der Straßen
- Verspätungen der S- und U-Bahnen
- Events in der Nähe
- Wetterdaten
Schnell stellte sich jedoch heraus, dass ich nicht alle diese Daten bekommen konnte, da sie nicht frei verfügbar sind bzw. es nicht gestattet ist, diese zu speichern. Da ich vorhatte, die gesammelten Daten auf GitHub zu speichern und verfügbar zu machen, war dies ein entscheidender Punkt, welche Daten in Frage kamen. Aus diesen Gründen fielen die Bahndaten vollkommen raus. Für die Straßenauslastung habe ich lediglich Daten für Köln gefunden und ich wollte es vermeiden, die Google API zu nutzen, da das durchaus seine eigenen Herausforderungen mit sich bringt. Es blieben also Event- und Wetterdaten.
Für die Wetterdaten des Deutschen Wetterdienstes kann das rdwd Packet genutzt werden. Da diese Daten bereits historisiert vorliegen, sind sie für diesen Blogbeitrag nebensächlich. Um an die verbleibenden Event- und Parkdaten zu kommen, haben sich die GitHub Actions als sehr nützlich erwiesen – auch wenn sie nicht ganz trivial in der Anwendung sind. Besonders der Umstand, dass diese kostenfrei genutzt werden können, machen sie zu einem empfehlenswerten Tool für solche Projekte.
Scrapen der Daten
Da sich dieser Beitrag nicht mit Details zum Thema Webscraping befassen wird, verweise ich an dieser Stelle auf den Beitrag von meinem Kollegen David.
Die Parkdaten stehen hier im XML-Format bereit und werden alle fünf Minuten aktualisiert. Sobald man die Struktur des XML verstanden hat, müsst ihr nur noch auf den richtigen Index zugreifen und ihr habt die Daten, die ihr möchtet.
In der Funktion get_parking_data() habe ich alles zusammengefasst, was ich benötige. Es wird ein Datensatz zur Area und ein Datensatz zu den einzelnen Parkhäusern erstellt.
Beispiel Datenauszug area
parkingAreaOccupancy;parkingAreaStatusTime;parkingAreaTotalNumberOfVacantParkingSpaces;
totalParkingCapacityLongTermOverride;totalParkingCapacityShortTermOverride;id;TIME
0.08401977;2021-12-01T01:07:00Z;556;150;607;1[Anlagenring];2021-12-01T01:07:02.720Z
0.31417114;2021-12-01T01:07:00Z;513;0;748;4[Bahnhofsviertel];2021-12-01T01:07:02.720Z
0.351417;2021-12-01T01:07:00Z;801;0;1235;5[Dom / Römer];2021-12-01T01:07:02.720Z
0.21266666;2021-12-01T01:07:00Z;1181;70;1500;2[Zeil];2021-12-01T01:07:02.720Z
Beispiel Datenauszug facility
parkingFacilityOccupancy;parkingFacilityStatus;parkingFacilityStatusTime;
totalNumberOfOccupiedParkingSpaces;totalNumberOfVacantParkingSpaces;
totalParkingCapacityLongTermOverride;totalParkingCapacityOverride;
totalParkingCapacityShortTermOverride;id;TIME
0.02;open;2021-12-01T01:02:00Z;4;196;150;350;200;24276[Turmcenter];2021-12-01T01:07:02.720Z
0.11547912;open;2021-12-01T01:02:00Z;47;360;0;407;407;18944[Alte Oper];2021-12-01T01:07:02.720Z
0.0027472528;open;2021-12-01T01:02:00Z;1;363;0;364;364;24281[Hauptbahnhof Süd];2021-12-01T01:07:02.720Z
0.609375;open;2021-12-01T01:02:00Z;234;150;0;384;384;105479[Baseler Platz];2021-12-01T01:07:02.720Z
Für die Eventdaten scrape ich die Seite stadtleben.de. Da es sich um eine HTML handelt, die recht gut strukturiert ist, kann ich über den Tag „kalenderListe“ auf die tabellarische Eventübersicht zugreifen. Das Resultat wird durch die Funktion get_event_data() erstellt.
Beispiel Datenauszug events
eventtitle;views;place;address;eventday;eventdate;request
Magical Sing Along - Das lustigste Mitsing-Event;12576;Bürgerhaus;64546 Mörfelden-Walldorf, Westendstraße 60;Freitag;2022-03-04;2022-03-04T02:24:14.234833Z
Velvet-Bar-Night;1460;Velvet Club;60311 Frankfurt, Weißfrauenstraße 12-16;Freitag;2022-03-04;2022-03-04T02:24:14.234833Z
Basta A-cappella-Band;465;Zeltpalast am Deutsche Bank Park;60528 Frankfurt am Main, Mörfelder Landstraße 362;Freitag;2022-03-04;2022-03-04T02:24:14.234833Z
BeThrifty Vintage Kilo Sale | Frankfurt | 04. & 05. …;1302;Batschkapp;60388 Frankfurt am Main, Gwinnerstraße 5;Freitag;2022-03-04;2022-03-04T02:24:14.234833Z
Automation der Abläufe – GitHub Actions
Das Grundgerüst steht. Ich habe je eine Funktion, die mir die Park- und Eventdaten beim Ausführen in eine .csv Datei schreiben. Da ich die Parkdaten alle fünf Minuten und die Eventdaten zur Sicherheit drei Mal am Tag abfragen möchte, kommen nun GitHub Actions ins Spiel.
Mit dieser Funktion von GitHub können neben Aktionen, die beim Mergen oder Committen auslösen, auch Workflows zeitlich geplant und durchgeführt werden. Hierfür wird eine .yml Datei im Order /.github/workflows erstellt.
Die Hauptbestandteile meines Workflows sind:
- Der
schedule– Alle zehn Minuten sollen die Funktionen ausgeführt werden. - Das OS – Da ich lokal auf einem Mac entwickle, nutze ich hier das
macOS-latest. - Umgebungsvariablen – Hier ist neben meinem GitHub Token auch der Pfad für das Paketmanagement
renventhalten - Die einzelnen
stepsim Workflow selbst
Der Workflow durchläuft die folgenden Schritte:
- Setup R
- Pakete laden mit renv
- Script ausführen um Daten zu scrapen
- Script ausführen um die README zu aktualisieren
- Pushen der neuen Daten zurück ins git
Jeder dieser Schritte ist an sich sehr klein und übersichtlich, jedoch liegt der Teufel wie so oft im Detail.
Limitation und Herausforderungen
Im Laufe der letzten Monate habe ich meinen Workflow immer wieder angepasst und optimiert, um den aufkommenden Fehlern und Problemen Herr zu werden. Nachfolgend also der Überblick über meine kondensierten Erfahrungen mit GitHub Actions.
Schedule Probleme
Wer zeitkritische Aktionen durchführen möchte, sollte auf andere Services zugreifen. GitHub Actions garantieren einem nicht, dass die Jobs exakt getimed werden (oder teilweise überhaupt durchgeführt werden). In der Tabelle sind die Zeiten zwischen zwei erfolgreichen Abfragen angegeben.
| Zeitspanne in Minuten | <= 5 | <= 10 | <= 20 | <= 60 | > 60 |
| Anzahl Abfragen | 1720 | 2049 | 5509 | 3023 | 194 |
Man sieht, dass die geplanten fünf Minuten Intervalle nicht immer eingehalten wurden. Hier sollte ich in Zukunft einen größeren Spielraum einplanen.
Merge Konflikte
Zu Beginn hatte ich zwei Workflows, einen für die Parkdaten und einen für die Events. Wenn diese sich zeitlich überlappt haben, dann kam es zu Merge-Konflikten, da beide Prozesse die README mit einen Zeitstempel updaten. Im Verlauf bin ich umgestiegen auf einen Workflow samt Errorhandling.
Auch wenn ein Durchlauf länger gedauert hat und der nächste bereits gestartet wurde, kam es beim Pushen zu Merge-Konflikten in den .csv-Daten. Lange Durchläufe entstanden häufig durch das R Setup und das Laden der packages. Als Konsequenz habe ich das Schedule-Intervall von fünf auf zehn Minuten erweitert.
Formatanpassungen
Es gab ein paar Situationen, in denen sich die Pfade oder Struktur der gescrapten Daten geändert haben, so dass ich meine Funktionen anpassen musste. Hierbei war die Einstellung, eine E-Mail zu bekommen, falls ein Prozess gescheitert ist, sehr hilfreich.
Fehlende Testmöglichkeiten
Es gibt bisher keine andere Möglichkeit ein Workflow-Script zu testen, als es wirklich laufen zu lassen. So kann man nach einem Tippfehler am Abend zu einer Mailflut mit gefailten Runs am Morgen aufwachen. Das sollte einen dennoch nicht davon abhalten einen lokalen Testlauf durchzuführen.
Kein Datenupdate
Seit Ende Dezember wurden die Parkdaten nicht mehr aktualisiert bzw. bereitgestellt. Das zeigt, dass selbst wenn man einen automatischen Prozess hat, man ihn dennoch weiter überwachen sollte. Ich habe dies erst später festgestellt, wodurch meine Abfragen Ende Dezember immer ins Leere liefen.
Fazit
Trotz der Komplikationen aus dem letzten Kapitel, empfinde ich das Ganze dennoch als einen massiven Erfolg. Während der letzten Monate habe ich mich immer wieder mit dem Thema befasst und die oben beschriebenen Tricks und Kniffe erlernt, die mir auch in Zukunft helfen werden, andere Probleme zu lösen. Ich hoffe, dass auch ihr ein paar wertvolle Hinweise mitnehmen und somit aus meinen Fehlern lernen könnt.
Da ich nun ein gutes halbes Jahr an Daten gesammelt haben, kann ich mich mit der Auswertung befassen. Das wird dann aber erst Gegenstand eines weiteren Blogbeitrages.
Einführung
Je komplexer ein beliebiges Data Science Projekt in Python wird, desto schwieriger wird es in der Regel, den Überblick darüber zu behalten, wie alle Module miteinander interagieren. Wenn man in einem Team an einem größeren Projekt arbeitet, wie es hier bei STATWORX oft der Fall ist, kann die Codebasis schnell so groß werden, dass die Komplexität abschreckend wirken kann. In einem typischen Szenario arbeitet jedes Teammitglied in seiner „Ecke“ des Projekts, so dass jeder nur über ein solides lokales Wissen über den Code des Projekts verfügt, aber möglicherweise nur eine vage Vorstellung von der Gesamtarchitektur des Projekts hat. Im Idealfall sollte jedoch jeder, der an dem Projekt beteiligt ist, einen guten globalen Überblick über das Projekt haben. Damit meine ich nicht, dass man wissen muss, wie jede Funktion intern funktioniert, sondern eher, dass man die Zuständigkeit der Hauptmodule kennt und weiß, wie sie miteinander verbunden sind.
Ein visuelles Hilfsmittel, um die globale Struktur kennenzulernen, kann ein Call Graph sein. Ein Call Graph ist ein gerichteter Graph, der anzeigt, welche Funktion welche Funktion aufruft. Er wird aus den Daten eines Python-Profilers wie cProfile erstellt.
Da sich ein solcher Graph in einem Projekt, an dem ich arbeite, als hilfreich erwiesen hat, habe ich ein Paket namens project_graph erstellt, das einen solchen Call Graph für ein beliebiges Python-Skript erstellt. Das Paket erstellt ein Profil des gegebenen Skripts über cProfile, konvertiert es in einen gefilterten Punktgraphen über gprof2dot und exportiert es schließlich als .png-Datei.
Warum sind Projektgrafiken nützlich?
Als erstes kleines Beispiel soll dieses einfache Modul dienen.
# test_script.py
import time
from tests.goodnight import sleep_five_seconds
def sleep_one_seconds():
time.sleep(1)
def sleep_two_seconds():
time.sleep(2)
for i in range(3):
sleep_one_seconds()
sleep_two_seconds()
sleep_five_seconds()Nach der Installation (siehe unten) wird durch Eingabe von project_graph test_script.py in die Kommandozeile die folgende png-Datei neben dem Skript platziert:
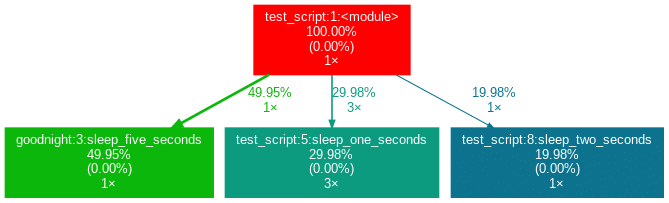
Das zu profilierende Skript dient immer als Ausgangspunkt und ist die Wurzel des Baums. Jedes Kästchen ist mit dem Namen einer Funktion, dem Gesamtprozentsatz der in der Funktion verbrachten Zeit und der Anzahl ihrer Aufrufe beschriftet. Die Zahl in Klammern gibt an, wieviel Zeit innerhalb einer Funktion verbracht wurde, jedoch ohne die Zeit in weiteren Unterfunktion zu berücksichtigen.
In diesem Fall wird die gesamte Zeit in der Funktion sleep des externen Moduls time verbracht, weshalb die Zahl 0,00% beträgt. In selbstgeschriebenen Funktionen wird nur selten viel Zeit verbracht, da die Arbeitslast eines Skripts in der Regel schnell auf sehr einfache Funktionen der Python-Implementierung selbst rausläuft. Neben den Pfeilen ist auch die Zeit angegeben, die eine Funktion an die andere weitergibt, zusammen mit der Anzahl der Aufrufe. Die Farben (ROT-GRÜN-BLAU, absteigend) und die Dicke der Pfeile zeigen die Relevanz der verschiedenen Stellen im Programm an.
Beachten Sie, dass sich die Prozentsätze der drei obigen Funktionen nicht zu 100 % aufaddieren. Der Grund dafür ist, dass der Graph so eingestellt ist, dass er nur selbst geschriebene Funktionen enthält. In diesem Fall hat das Importieren des Moduls time den Python-Interpreter dazu veranlasst, 0,04% der Zeit für eine Funktion des Moduls importlib aufzuwenden.
Auswertung mit externen Packages
Betrachten wir ein zweites Beispiel:
# test_script_2.py
import pandas as pd
from tests.goodnight import sleep_five_seconds
# some random madness
for i in range(1000):
a_frame = pd.DataFrame([[1,2,3]])
sleep_five_seconds()In diesem Skript wird ein Teil der Arbeit in einem externen Paket erledigt, das auf der Top-Ebene und nicht in einer benutzerdefinierten Funktion aufgerufen wird. Um dies im Graphen zu erfassen, können wir das externe Paket (pandas) mit der Flag -x hinzufügen. Die Initialisierung eines Pandas DataFrame wird jedoch in vielen Pandas-internen Funktionen durchgeführt. Offen gesagt, bin ich persönlich nicht an den inneren Verwicklungen von pandas interessiert, weshalb ich möchte, dass der Baum nicht zu tief in die Pandas-Mechanik „hineinwächst“. Diesem Umstand kann man Rechnung tragen, indem man nur Funktionen auftauchen lässt, die einen minimalen Prozentsatz der Laufzeit in ihnen verbringen. Genau dies kann mit der -m-Flag erreicht werden.
In Kombination ergibt project_graph -m 8 -x pandas test_script_2.py das folgende Ergebnis:
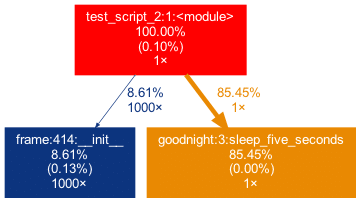
Spaß(-Beispiele) beiseite, nun wollen wir uns ernsteren Dingen zuwenden. Ein echtes Data Science Projekt könnte wie dieses aussehen:
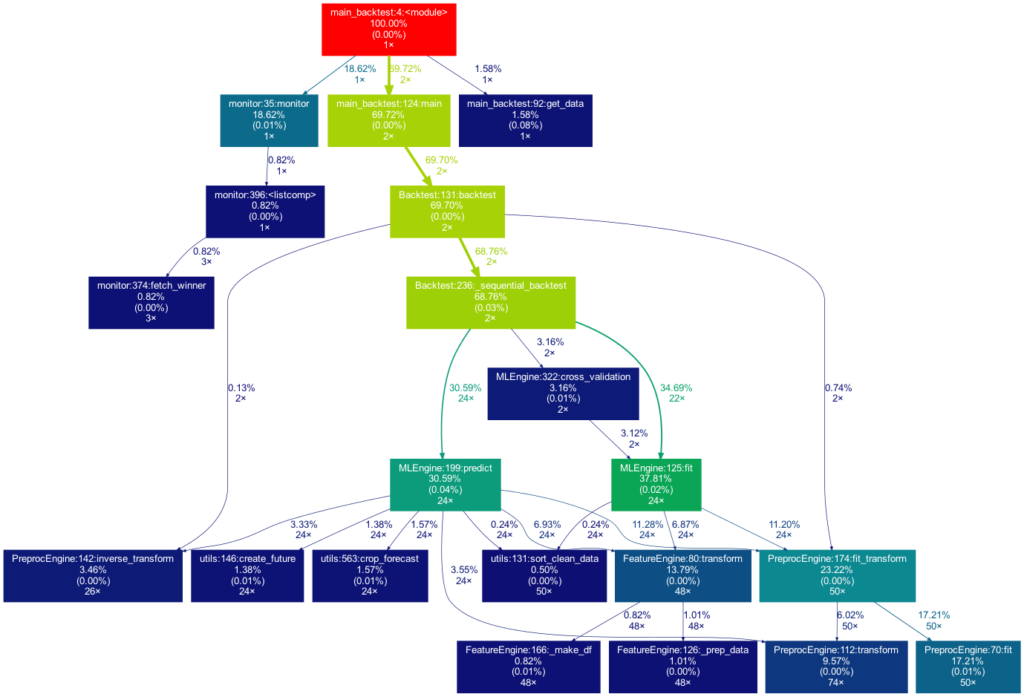
Dieses Mal ist der Baum viel größer. Er ist sogar noch größer als in der Abbildung zu sehen, da viel mehr selbst geschriebene Funktionen aufgerufen werden. Sie werden jedoch aus Gründen der Übersichtlichkeit aus dem Baum entfernt, da Funktionen, für die weniger als 0,5 % der Gesamtzeit aufgewendet werden, herausgefiltert werden (dies ist die Standardeinstellung für die -m Flag). Beachten Sie, dass ein solches Diagramm auch bei der Suche nach Leistungsengpässen sehr vorteilhaft ist. Man sieht sofort, welche Funktionen den größten Teil der Arbeitslast tragen, wann sie aufgerufen werden und wie oft sie aufgerufen werden. Das kann Sie davor bewahren, Ihr Programm an den falschen Stellen zu optimieren und dabei den Elefanten im Raum zu übersehen.
Wie man project graph verwendet
Installation
Gehen Sie in Ihrer Projektumgebung wie folgt vor:
brew install graphviz
pip install git+https://github.com/fior-di-latte/project_graph.gitVerwendung
Wechseln Sie in der Projektumgebung in das aktuelle Arbeitsverzeichnis des Projekts (das ist wichtig!) und geben Sie für die Standardverwendung ein:
project_graph myscript.pyWenn Ihr Skript einen argparser enthält, verwenden Sie (vergessen Sie nicht die Anführungsstriche!):
project_graph "myscript.py <arg1> <arg2> (...)"Wenn Sie den gesamten Graphen sehen wollen, einschließlich aller externen Pakete, verwenden Sie:
project_graph -a myscript.pyWenn Sie eine andere Sichtbarkeitsschwelle als 1% verwenden wollen, benutzen Sie:
project_graph -m <percent_value> myscript.pyWenn Sie schließlich externe Pakete in den Graphen aufnehmen wollen, können Sie sie wie folgt angeben:
project_graph -x <package1> -x <package2> (...) myscript.pySchluss & Hinweise
Dieses Paket hat einige Schwächen, von denen die meisten behoben werden können, z.B. durch Formatierung des Codes in einen funktionsbasierten Stil, durch Trimmen mit der -m-Flag oder durch Hinzufügen von Paketen mit der-x-Flag. Wenn etwas seltsam erscheint ist der erste Schritt wahrscheinlich die Verwendung der -a-Flag zur Fehlersuche. Wesentliche Einschränkungen sind die folgenden:
- Es funktioniert nur auf Unix-Systemen.
- Es zeigt keinen wahrheitsgetreuen Graphen an, wenn es mit Multiprocessing verwendet wird. Der Grund dafür ist, dass cProfile nicht mit Multiprocessing kompatibel ist. Wenn Multiprocessing verwendet wird, wird nur der Root-Prozess profiliert, was zu falschen Berechnungszeiten im Graphen führt. Wechseln Sie zu einer nicht-parallelen Version des Zielskripts.
- Die Profilerstellung eines Skripts kann zu einem beträchtlichen Overhead bei der Berechnung führen. Es kann sinnvoll sein, die in Ihrem Skript geleistete Arbeit zu verringern (d. h. die Menge der Eingabedaten zu reduzieren). In diesem Fall kann die in den Funktionen verbrachte Zeit natürlich massiv verzerrt werden, wenn die Funktionen nicht linear skalieren.
- Verschachtelte Funktionen werden im Diagramm nicht angezeigt. Insbesondere ein Dekorator verschachtelt implizit Ihre Funktion und versteckt sie daher. Das heißt, wenn Sie einen externen Dekorator verwenden, vergessen Sie nicht, das Paket des Dekorators über die
-xFlag hinzuzufügen (zum Beispielproject_graph -x numba myscript.py). - Wenn Ihre selbst geschriebene Funktion ausschließlich von einer Funktion eines externen Pakets aufgerufen wird, müssen Sie das externe Paket manuell mit der
-xFlag hinzufügen. Andernfalls wird Ihre Funktion nicht im Baum auftauchen, da ihr Parent eine externe Funktion ist und daher nicht berücksichtigt wird.
Sie können das kleine Paket gerne für Ihr eigenes Projekt verwenden, sei es für Leistungsanalysen, Code-Einführungen für neue Teammitglieder oder aus reiner Neugier. Was mich betrifft, so finde ich es sehr befriedigend, eine solche Visualisierung meiner Projekte zu sehen. Wenn Sie Probleme bei der Verwendung haben, zögern Sie nicht, mich auf Github zu kontaktieren (https://github.com/fior-di-latte/project_graph/).
PS: Wenn Sie nach einem ähnlichen Paket in R suchen, sehen Sie sich Jakobs Beitrag über Flussdiagramme von Funktionen an.
Du willst Python lernen? Oder bist du ein R-Profi und dir entfallen bei der Arbeit mit Python regelmäßig die wichtigen Funktionen und Befehle? Oder vielleicht brauchst du von Zeit zu Zeit eine kleine Gedächtnisstütze beim Programmieren? Genau dafür wurden Cheatsheets erfunden!
Cheatsheets helfen dir in all diesen Situationen weiter. Unser erstes Cheatsheet mit den Grundlagen von Python ist der Start einer neuen Blog-Serie, in der weitere Cheatsheets in unserem einzigartigen STATWORX Stil folgen werden.
Du kannst also neugierig sein auf unsere Serie von neuen Python-Cheatsheets, die sowohl Grundlagen als auch Pakete und Arbeitsfelder, die für Data Science relevant sind, behandeln werden.
Unsere Cheatsheets stehen euch zum kostenfreien Download frei zur Verfügung, ohne Anmeldung oder sonstige Paywall.
Warum haben wir neue Cheatsheets erstellt?
Als erfahrene R User sucht man schier endlos nach entsprechend modernen Python Cheatsheets, ähnlich denen, die du von R Studio kennst.
Klar, es gibt eine Vielzahl von Cheatsheets für jeden Themenbereich, die sich aber in Design und Inhalt stark unterscheiden. Sobald man mehrere Cheatsheets in unterschiedlichen Designs verwendet, muss man sich ständig neu orientieren und verliert so insgesamt viel Zeit. Für uns als Data Scientists ist es wichtig, einheitliche Cheatsheets zu haben, anhand derer wir schnell die gewünschte Funktion oder den Befehl finden können.
Diesem nervigen Zusammensuchen von Informationen wollen wir entgegenwirken. Daher möchten wir auf unserem Blog zukünftig regelmäßig neue Cheatsheets in einer Designsprache veröffentlichen – und euch alle an dieser Arbeitserleichterung teilhaben lassen.
Was enthält das erste Cheatsheet?
Unser erstes Cheatsheet in dieser Reihe richtet sich in erster Linie an Python-Neulinge, an R-Nutzer, die Python seltener verwenden, oder an Leute, die gerade erst anfangen, mit Python zu arbeiten.
Es erleichtert den Einstieg und Überblick in Python. Die grundlegende Syntax, die Datentypen und der Umgang mit diesen werden vorgestellt und grundlegende Kontrollstrukturen eingeführt. So kannst du schnell auf die Inhalte zugreifen, die du z.B. in unserer STATWORX Academy gelernt hast oder dir die Grundlagen für dein nächstes Programmierprojekt ins Gedächtnis rufen.
Was behandelt das STATWORX Cheatsheet Episode 2?
Das nächste Cheatsheet behandelt den ersten Schritt eines Data Scientists in einem neuen Projekt: Data Wrangling. Außerdem erwartet dich ein Cheatsheet für pandas über das Laden, Auswählen, Manipulieren, Aggregieren und Zusammenführen von Daten. Happy Coding!
Im ersten Beitrag dieser Serie haben wir Transfer Learning im Detail besprochen und ein Modell zur Klassifizierung von Automodellen erstellt. In diesem Beitrag werden wir das Problem der Modellbereitstellung am Beispiel des im ersten Beitrags vorgestellten TransferModel diskutieren.
Ein Modell ist in der Praxis nutzlos, wenn es keine einfache Möglichkeit gibt, damit zu interagieren. Mit anderen Worten: Wir brauchen eine API für unsere Modelle. TensorFlow Serving wurde entwickelt, um diese Funktionalitäten für TensorFlow-Modelle bereitzustellen. In diesem Beitrag zeigen wir, wie ein TensorFlow Serving Server in einem Docker-Container gestartet werden kann und wie wir mit dem Server über HTTP-Anfragen interagieren können.
Wenn ihr noch nie mit Docker gearbeitet habt, empfehlen wir, dieses Tutorial von Docker durchzuarbeiten, bevor ihr diesen Artikel lest. Wenn ihr euch ein Beispiel für das Deployment in Docker ansehen möchtet, empfehlen wir euch, diesen Blogbeitrag von unserem Kollegen Oliver Guggenbühl zu lesen, in dem beschrieben wird, wie ein R-Skript in Docker ausgeführt werden kann.
Einführung in TensorFlow Serving
Zum Einstieg geben wir euch zunächst einen Überblick über TensorFlow Serving.
TensorFlow Serving ist das Serving-System von TensorFlow, das entwickelt wurde, um das Deployment von verschiedenen Modellen mit einer einheitlichen API zu ermöglichen. Unter Verwendung der Abstraktion von Servables, die im Grunde Objekte sind, mit denen Inferenz durchgeführt werden kann, ist es möglich, mehrere Versionen von deployten Modellen zu serven. Das ermöglicht zum Beispiel, dass eine neue Version eines Modells hochgeladen werden kann, während die vorherige Version noch für Kunden verfügbar ist. Im Großen und Ganzen sind sogenannte Manager für die Verwaltung des Lebenszyklus von Servables verantwortlich, d. h. für das Laden, Bereitstellen und Löschen.
In diesem Beitrag werden wir zeigen, wie eine einzelne Modellversion deployed werden kann. Die unten aufgeführten Code-Beispiele zeigen, wie ein Server in einem Docker-Container gestartet werden kann und wie die Predict API verwendet werden kann, um mit dem Modell zu interagieren. Um mehr über TensorFlow Serving zu erfahren, verweisen wir auf die TensorFlow-Website.
Implementierung
Wir werden nun die folgenden drei Schritte besprechen, die erforderlich sind, um das Modell einzusetzen und Requests zu senden.
- Speichern eines Modells im richtigen Format und in der richtigen Ordnerstruktur mit TensorFlow SavedModel
- Ausführen eines Serving-Servers innerhalb eines Docker-Containers
- Interaktion mit dem Modell über REST Requests
Speichern von TensorFlow-Modellen
Für diejenigen, die den ersten Beitrag dieser Serie nicht gelesen haben, folgt nun eine kurze Zusammenfassung der wichtigsten Punkte, die zum Verständnis des nachfolgenden Codes notwendig sind:
Das TransferModel.model (unten im Code auch self.model) ist eine tf.keras.Model Instanz, also kann es mit der eingebauten save Methode gespeichert werden. Da das Modell auf im Internet gescrapten Daten trainiert wurde, können sich die Klassenbezeichnungen beim erneuten Scraping der Daten ändern. Wir speichern daher die Index-Klassen-Zuordnung beim Speichern des Modells in classes.pickle ab. TensorFlow Serving erfordert, dass das Modell im SavedModel Format gespeichert wird. Wenn Sie tf.keras.Model.save verwenden, muss der Pfad ein Ordnername sein, sonst wird das Modell in einem anderen, inkompatiblen Format (z.B. HDF5) gespeichert. Im Code unten enthält folderpath den Pfad des Ordners, in dem wir alle modellrelevanten Informationen speichern wollen. Das SavedModel wird unter folderpath/model gespeichert und das Class Mapping wird als folderpath/classes.pickle gespeichert.
def save(self, folderpath: str):
"""
Save the model using tf.keras.model.save
Args:
folderpath: (Full) Path to folder where model should be stored
"""
# Make sure folderpath ends on slash, else fix
if not folderpath.endswith("/"):
folderpath += "/"
if self.model is not None:
os.mkdir(folderpath)
model_path = folderpath + "model"
# Save model to model dir
self.model.save(filepath=model_path)
# Save associated class mapping
class_df = pd.DataFrame({'classes': self.classes})
class_df.to_pickle(folderpath + "classes.pickle")
else:
raise AttributeError('Model does not exist')TensorFlow Serving im Docker Container starten
Nachdem wir das Modell auf der Festplatte gespeichert haben, müssen wir nun den TensorFlow Serving Server starten. Am schnellsten deployen kann man TensorFlow Serving mithilfe eines Docker-Containers. Der erste Schritt ist daher das Ziehen des TensorFlow Serving Images von DockerHub. Das kann im Terminal mit dem Befehl docker pull tensorflow/serving gemacht werden.
Dann können wir den unten stehenden Code verwenden, um einen TensorFlow Serving Container zu starten. Er führt den Shell-Befehl zum Starten eines Containers aus. Die in docker_run_cmd gesetzten Optionen sind die folgenden:
- Das Serving-Image exponiert Port 8501 für die REST-API, die wir später zum Senden von Anfragen verwenden werden. Wir mappen mithilfe der
-p– Flag also den Host-Port 8501 auf den Port 8501 des Containers. - Als nächstes binden wir unser Modell mit
-vin den Container ein. Es ist wichtig, dass das Modell in einem versionierten Ordner gespeichert ist (hier MODEL_VERSION=1); andernfalls wird das Serving-Image das Modell nicht finden. Dermodel_path_guestmuss also die Form<path>/<model name>/MODEL_VERSIONhaben, wobeiMODEL_VERSIONeine ganze Zahl ist. - Mit
-ekönnen wir die UmgebungsvariableMODEL_NAMEsetzen, die den Namen unseres Modells enthält. - Die Option
--name tf_servingwird nur benötigt, um unserem neuen Docker-Container einen bestimmten Namen zuzuweisen.
Wenn wir versuchen, diese Datei zweimal hintereinander auszuführen, wird der Docker-Befehl beim zweiten Mal nicht ausgeführt, da bereits ein Container mit dem Namen tf_serving existiert. Um dieses Problem zu vermeiden, verwenden wir docker_run_cmd_cond. Hier prüfen wir zunächst, ob ein Container mit diesem spezifischen Namen bereits existiert und läuft. Wenn ja, lassen wir ihn gleich; wenn nicht, prüfen wir, ob eine beendete Version des Containers existiert. Wenn ja, wird diese gelöscht und ein neuer Container gestartet; wenn nicht, wird direkt ein neuer Container erstellt.
import os
MODEL_FOLDER = 'models'
MODEL_SAVED_NAME = 'resnet_unfreeze_all_filtered.tf'
MODEL_NAME = 'resnet_unfreeze_all_filtered'
MODEL_VERSION = '1'
# Define paths on host and guest system
model_path_host = os.path.join(os.getcwd(), MODEL_FOLDER, MODEL_SAVED_NAME, 'model')
model_path_guest = os.path.join('/models', MODEL_NAME, MODEL_VERSION)
# Container start command
docker_run_cmd = f'docker run '
f'-p 8501:8501 '
f'-v {model_path_host}:{model_path_guest} '
f'-e MODEL_NAME={MODEL_NAME} '
f'-d '
f'--name tf_serving '
f'tensorflow/serving'
# If container is not running, create a new instance and run it
docker_run_cmd_cond = f'if [ ! "![Rendered by QuickLaTeX.com (docker ps -q -f name=tf_serving)" ]; then n' f' if [ "](https://www.statworx.com/wp-content/ql-cache/quicklatex.com-1aec58f1795a0cc6ff2d2f7d44be6a12_l3.png) (docker ps -aq -f status=exited -f name=tf_serving)" ]; then n'
f' docker rm tf_serving n'
f' fi n'
f' {docker_run_cmd} n'
f'fi'
# Start container
os.system(docker_run_cmd_cond)
(docker ps -aq -f status=exited -f name=tf_serving)" ]; then n'
f' docker rm tf_serving n'
f' fi n'
f' {docker_run_cmd} n'
f'fi'
# Start container
os.system(docker_run_cmd_cond)Anstatt das Modell von unserer lokalen Festplatte zu mounten, indem wir das -v-Flag im Docker-Befehl verwenden, könnten wir das Modell auch in das Docker-Image kopieren, so dass das Modell einfach durch das Ausführen eines Containers und die Angabe der Port-Zuweisungen bedient werden könnte. Es ist wichtig zu beachten, dass in diesem Fall das Modell mit der Ordnerstruktur Ordnerpfad/<Modellname>/1 gespeichert werden muss, wie oben erklärt. Wenn dies nicht der Fall ist, wird der TensorFlow Serving Container das Modell nicht finden. Wir werden hier nicht weiter auf diesen Fall eingehen. Wenn ihr daran interessiert seid, eure Modelle auf diese Weise zu deployen, verweisen wir auf diese Anleitung auf der TensorFlow Webseite.
REST Request
Da das Modell nun geserved ist und bereit zur Verwendung ist, brauchen wir einen Weg, um damit zu interagieren. TensorFlow Serving bietet zwei Optionen, um Anfragen an den Server zu senden: gRCP und REST API, welche beide an unterschiedlichen Ports verfügbar sind. Im folgenden Codebeispiel werden wir REST verwenden, um das Modell abzufragen.
Zuerst laden wir ein Bild von der Festplatte, für das wir eine Vorhersage machen wollen. Dies kann mit dem image Modul von TensorFlow gemacht werden. Als nächstes konvertieren wir das Bild in ein Numpy-Array mittels der img_to_array-Methode. Der nächste und letzte Schritt ist entscheidend für unseren Car Classifier Use Case: da wir das Trainingsbild vorverarbeitet haben, bevor wir unser Modell trainiert haben (z.B. Normalisierung), müssen wir die gleiche Transformation auf das Bild anwenden, das wir vorhersagen wollen. Die praktische Funktion „preprocess_input“ sorgt dafür, dass alle notwendigen Transformationen auf unser Bild angewendet werden.
from tensorflow.keras.preprocessing import image
from tensorflow.keras.applications.resnet_v2 import preprocess_input
# Load image
img = image.load_img(path, target_size=(224, 224))
img = image.img_to_array(img)
# Preprocess and reshape data
img = preprocess_input(img)
img = img.reshape(-1, *img.shape)Die RESTful API von TensorFlow Serving bietet mehrere Endpunkte. Im Allgemeinen akzeptiert die API Post-Requests der folgenden Struktur:
POST http://host:port/<URI>:<VERB>
URI: /v1/models/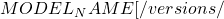 {MODEL_VERSION}]
VERB: classify|regress|predict
{MODEL_VERSION}]
VERB: classify|regress|predictFür unser Modell können wir die folgende URL für Vorhersagen verwenden: http://localhost:8501/v1/models/resnet_unfreeze_all_filtered:predict
Die Portnummer (hier 8501) ist der Port des Hosts, den wir oben angegeben haben, um ihn auf den Port 8501 des Serving-Images abzubilden. Wie oben erwähnt, ist 8501 der Port des Serving-Containers, der für die REST-API verwendet wird. Die Modellversion ist optional und wird standardmäßig auf die neueste Version gesetzt, wenn sie weggelassen wird.
In Python kann die Bibliothek requests verwendet werden, um HTTP-Anfragen zu senden. Wie in der Dokumentation angegeben, muss der Request-Body für die predict API ein JSON-Objekt mit den unten aufgeführten Schlüssel-Wert-Paaren sein:
signature_name– zu verwendende Signatur (weitere Informationen finden Sie in der Dokumentation)instances– Modelleingabe im Zeilenformat
import json
import requests
# Send image as list to TF serving via json dump
request_url = 'http://localhost:8501/v1/models/resnet_unfreeze_all_filtered:predict'
request_body = json.dumps({"signature_name": "serving_default", "instances": img.tolist()})
request_headers = {"content-type": "application/json"}
json_response = requests.post(request_url, data=request_body, headers=request_headers)
response_body = json.loads(json_response.text)
predictions = response_body['predictions']
# Get label from prediction
y_hat_idx = np.argmax(predictions)
y_hat = classes[y_hat_idx]Der Response-Body ist ebenfalls ein JSON-Objekt mit einem einzigen Schlüssel namens predictions. Da wir für jede Zeile in den Instanzen die Wahrscheinlichkeit für alle 300 Klassen erhalten, verwenden wir np.argmax, um die wahrscheinlichste Klasse zurückzugeben. Alternativ hätten wir auch die übergeordnete classify-API verwenden können.
Fazit
In diesem zweiten Blog-Artikel der Serie „Car Model Classification“ haben wir gelernt, wie ein TensorFlow-Modell zur Bilderkennung mittels TensorFlow Serving als RestAPI bereitgestellt werden kann, und wie damit Modellabfragen ausgeführt werden können.
Dazu haben wir zuerst das Modell im SavedModel Format abgespeichert. Als nächstes haben wir den TensorFlow Serving-Server in einem Docker-Container gestartet. Schließlich haben wir gezeigt, wie man Vorhersagen aus dem Modell mit Hilfe der API-Endpunkte und einem korrekt spezifizierten Request Body anfordert.
Ein Hauptkritikpunkt an Deep Learning Modellen jeglicher Art ist die fehlende Erklärbarkeit der Vorhersagen. Im dritten Beitrag werden wir zeigen, wie man Modellvorhersagen mit einer Methode namens Grad-CAM erklären kann.
Deep Learning ist eines der Themen im Bereich der künstlichen Intelligenz, die uns bei STATWORX besonders faszinieren. In dieser Blogserie möchten wir veranschaulichen, wie ein End-to-end Deep Learning Projekt implementiert werden kann. Dabei verwenden wir die TensorFlow 2.x Bibliothek für die Implementierung.
Die Themen der 4-teiligen Blogserie umfassen:
- Transfer Learning für Computer Vision
- Deployment über TensorFlow Serving
- Interpretierbarkeit von Deep-Learning-Modellen mittels Grad-CAM
- Integration des Modells in ein Dashboard
Im ersten Teil zeigen wir, wie man Transfer Learning nutzen kann, um die Marke eines Autos mittels Bildklassifizierung vorherzusagen. Wir beginnen mit einem kurzen Überblick über Transfer Learning und das ResNet und gehen dann auf die Details der Implementierung ein. Der vorgestellte Code ist in diesem Github Repository zu finden.
Einführung: Transfer Learning & ResNet
Was ist Transfer Learning?
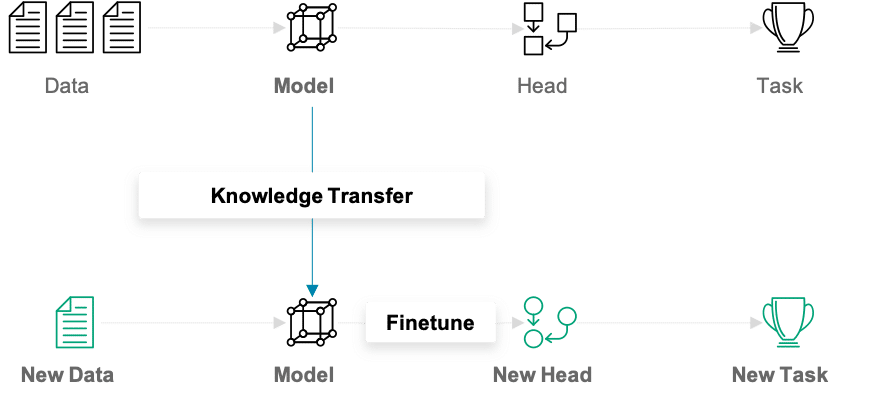
Beim traditionellen (Machine) Learning entwickeln wir ein Modell und trainieren es auf neuen Daten für jede neue Aufgabe, die ansteht. Transfer Learning unterscheidet sich von diesem Ansatz dadurch, dass das gesammelte Wissen von einer Aufgabe auf eine andere übertragen wird. Dieser Ansatz ist besonders nützlich, wenn einem zu wenige Trainingsdaten zur Verfügung stehen. Modelle, die für ein ähnliches Problem vortrainiert wurden, können als Ausgangspunkt für das Training neuer Modelle verwendet werden. Die vortrainierten Modelle werden als Basismodelle bezeichnet.
In unserem Beispiel kann ein Deep Learning-Modell, das auf dem ImageNet-Datensatz trainiert wurde, als Ausgangspunkt für die Erstellung eines Klassifikationsnetzwerks für Automodelle verwendet werden. Die Hauptidee hinter dem Transfer Learning für Deep Learning-Modelle ist, dass die ersten Layer eines Netzwerks verwendet werden, um wichtige High-Level-Features zu extrahieren, die für die jeweilige Art der behandelten Daten ähnlich bleiben. Die finalen Layer, auch „head“ genannt, des ursprünglichen Netzwerks werden durch einen benutzerdefinierten head ersetzt, der für das vorliegende Problem geeignet ist. Die Gewichte im head werden zufällig initialisiert, und das resultierende Netz kann für die spezifische Aufgabe trainiert werden.
Es gibt verschiedene Möglichkeiten, wie das Basismodell beim Training behandelt werden kann. Im ersten Schritt können seine Gewichte fixiert werden. Wenn der Lernfortschritt darauf schließen lässt, dass das Modell nicht flexibel genug ist, können bestimmte Layer oder das gesamte Basismodell auch mit trainiert werden. Ein weiterer wichtiger Aspekt, den es zu beachten gilt, ist, dass der Input die gleiche Dimensionalität haben muss wie die Daten, auf denen das Basismodell initial trainiert wurde – sofern die ersten Layer des Basismodells festgehalten werden sollen.
Als nächstes stellen wir kurz das ResNet vor, eine beliebte und leistungsfähige CNN-Architektur für Bilddaten. Anschließend zeigen wir, wie wir Transfer Learning mit ResNet zur Klassifizierung von Automodellen eingesetzt haben.
Was ist ResNet?
Das Training von Deep Neural Networks kann aufgrund des sogenannten Vanishing Gradient-Problems schnell zur Herausforderung werden. Aber was sind Vanishing Gradients? Neuronale Netze werden in der Regel mit Back-Propagation trainiert. Dieser Algorithmus nutzt die Kettenregel der Differentialrechnung, um Gradienten in tieferen Layern des Netzes abzuleiten, indem Gradienten aus früheren Layern multipliziert werden. Da Gradienten in Deep Networks wiederholt multipliziert werden, können sie sich während der Backpropagation schnell infinitesimal kleinen Werten annähern.
ResNet ist ein CNN-Netz, welches das Problem des Vanishing Gradients mit sogenannten Residualblöcken löst (eine gute Erklärung, warum sie ‚Residual‘ heißen, findest du hier). Im Residualblock wird die unmodifizierte Eingabe an das nächste Layer weitergereicht, indem sie zum Ausgang eines Layers addiert wird (siehe Abbildung rechts). Diese Modifikation sorgt dafür, dass ein besserer Informationsfluss von der Eingabe zu den tieferen Layers möglich ist. Die gesamte ResNet-Architektur ist im rechten Netzwerk in der linken Abbildung unten dargestellt. Weiter sind daneben ein klassisches CNN und das VGG-19-Netzwerk, eine weitere Standard-CNN-Architektur, abgebildet.
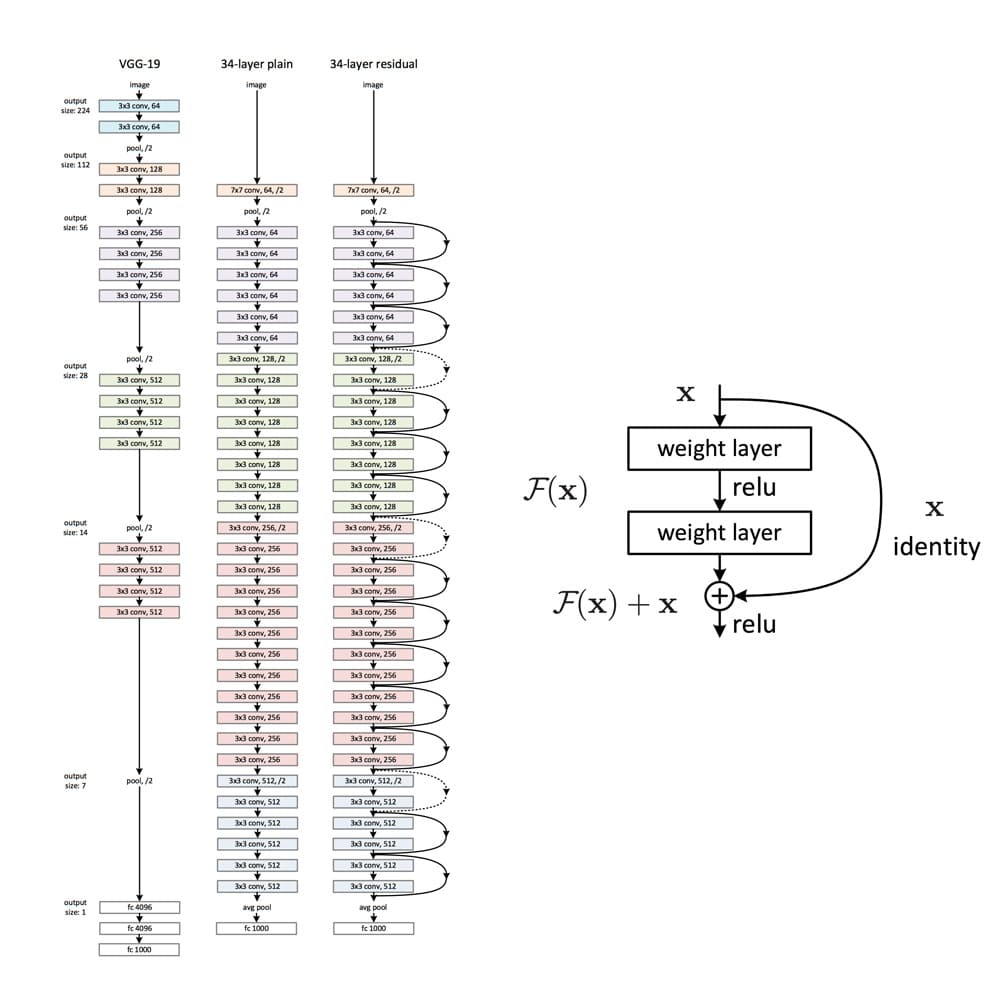
ResNet hat sich als leistungsfähige Netzarchitektur für Bildklassifikationsprobleme erwiesen. Zum Beispiel hat ein Ensemble von ResNets mit 152 Layern den ILSVRC 2015 Bildklassifikationswettbewerb gewonnen. Im Modul tensorflow.keras.application sind vortrainierte ResNet-Modelle unterschiedlicher Größe verfügbar, nämlich ResNet50, ResNet101, ResNet152 und die entsprechenden zweiten Versionen (ResNet50V2, …). Die Zahl hinter dem Modellnamen gibt die Anzahl der Layer an, über die die Netze verfügen. Die verfügbaren Gewichte sind auf dem ImageNet-Datensatz vortrainiert. Die Modelle wurden auf großen Rechenclustern unter Verwendung von spezialisierter Hardware (z.B. TPU) über signifikante Zeiträume trainiert. Transfer Learning ermöglicht es uns daher, diese Trainingsergebnisse zu nutzen und die erhaltenen Gewichte als Ausgangspunkt zu verwenden.
Klassifizierung von Automodellen
Als anschauliches Beispiel für die Anwendung von Transfer Learning behandeln wir das Problem der Klassifizierung des Automodells anhand eines Bildes des Autos. Wir beginnen mit der Beschreibung des verwendeten Datensatzes und wie wir unerwünschte Beispiele aus dem Datensatz herausfiltern können. Anschließend gehen wir darauf ein, wie eine Datenpipeline mit tensorflow.data eingerichtet werden kann. Im zweiten Abschnitt werden wir die Modellimplementierung durchgehen und aufzeigen, auf welche Aspekte ihr beim Training und bei der Inferenz besonders achten müsst.
Datenvorbereitung
Wir haben den Datensatz aus diesem GitHub Repo verwendet – dort könnt ihr den gesamten Datensatz herunterladen. Der Autor hat einen Datascraper gebaut, um alle Autobilder von der Car Connection Website zu scrapen. Er erklärt, dass viele Bilder aus dem Innenraum der Autos stammen. Da sie im Datensatz nicht erwünscht sind, filtern wir sie anhand der Pixelfarbe heraus. Der Datensatz enthält 64’467 jpg-Bilder, wobei die Dateinamen Informationen über die Automarke, das Modell, das Baujahr usw. enthalten. Für einen detaillierteren Einblick in den Datensatz empfehlen wir euch, das originale GitHub Repo zu konsultieren. Hier sind drei Beispielbilder:

Bei der Betrachtung der Daten haben wir festgestellt, dass im Datensatz noch viele unerwünschte Bilder enthalten waren, z.B. Bilder von Außenspiegeln, Türgriffen, GPS-Panels oder Leuchten. Beispiele für unerwünschte Bilder sind hier zu sehen:

Daher ist es von Vorteil, die Daten zusätzlich vorzufiltern, um mehr unerwünschte Bilder zu entfernen.
Filtern unerwünschter Bilder aus dem Datensatz
Es gibt mehrere mögliche Ansätze, um Nicht-Auto-Bilder aus dem Datensatz herauszufiltern:
- Verwendung eines vortrainierten Modells
- Ein anderes Modell trainieren, um Auto/Nicht-Auto zu klassifizieren
- Trainieren eines Generative Networks auf einem Auto-Datensatz und Verwendung des Diskriminatorteil des Netzwerks
Wir haben uns für den ersten Ansatz entschieden, da er der direkteste ist und ausgezeichnete, vortrainierte Modelle leicht verfügbar sind. Wenn ihr den zweiten oder dritten Ansatz verfolgen wollt, könnt ihr z. B. diesen Datensatz verwenden, um das Modell zu trainieren. Dieser Datensatz enthält nur Bilder von Autos, ist aber deutlich kleiner als der von uns verwendete Datensatz.
Unsere Wahl fiel auf das ResNet50V2 im Modul tensorflow.keras.applications mit den vortrainierten „imagenet“-Gewichten. In einem ersten Schritt müssen wir jetzt die Indizes und Klassennamen der imagenet-Labels herausfinden, die den Autobildern entsprechen.
# Class labels in imagenet corresponding to cars
CAR_IDX = [656, 627, 817, 511, 468, 751, 705, 757, 717, 734, 654, 675, 864, 609, 436]
CAR_CLASSES = ['minivan', 'limousine', 'sports_car', 'convertible', 'cab', 'racer', 'passenger_car', 'recreational_vehicle', 'pickup', 'police_van', 'minibus', 'moving_van', 'tow_truck', 'jeep', 'landrover', 'beach_wagon']Als nächstes laden wir das vortrainierte ResNet50V2-Modell.
from tensorflow.keras.applications import ResNet50V2
model = ResNet50V2(weights='imagenet')Wir können dieses Modell nun verwenden, um die Bilder zu klassifizieren. Die Bilder, die der Vorhersagemethode zugeführt werden, müssen identisch skaliert sein wie die Bilder, die zum Training verwendet wurden. Die verschiedenen ResNet-Modelle werden auf unterschiedlich skalierten Bilddaten trainiert. Es ist daher wichtig, das richtige Preprocessing anzuwenden.
from tensorflow.keras.applications.resnet_v2 import preprocess_input
image = tf.io.read_file(filename)
image = tf.image.decode_jpeg(image)
image = tf.cast(image, tf.float32)
image = tf.image.resize_with_crop_or_pad(image, target_height=224, target_width=224)
image = preprocess_input(image)
predictions = model.predict(image)Es gibt verschiedene Ideen, wie die erhaltenen Vorhersagen für die Autoerkennung verwendet werden können.
- Ist eine der
CAR_CLASSESunter den Top-k-Vorhersagen? - Ist die kumulierte Wahrscheinlichkeit der
CAR_CLASSESin den Vorhersagen größer als ein definierter Schwellenwert? - Spezielle Behandlung unerwünschter Bilder (z. B. Erkennen und Herausfiltern von Rädern)?
Wir zeigen den Code für den Vergleich der kumulierten Wahrscheinlichkeitsmaße über die CAR_CLASSES.
def is_car_acc_prob(predictions, thresh=THRESH, car_idx=CAR_IDX):
"""
Determine if car on image by accumulating probabilities of car prediction and comparing to threshold
Args:
predictions: (?, 1000) matrix of probability predictions resulting from ResNet with imagenet weights
thresh: threshold accumulative probability over which an image is considered a car
car_idx: indices corresponding to cars
Returns:
np.array of booleans describing if car or not
"""
predictions = np.array(predictions, dtype=float)
car_probs = predictions[:, car_idx]
car_probs_acc = car_probs.sum(axis=1)
return car_probs_acc > threshJe höher der Schwellenwert eingestellt ist, desto strenger ist das Filterverfahren. Ein Wert für den Schwellenwert, der gute Ergebnisse liefert, ist THRESH = 0.1. Damit wird sichergestellt, dass nicht zu viele echte Bilder von Autos verloren gehen. Die Wahl eines geeigneten Schwellenwerts bleibt jedoch eine subjektive Angelegenheit.
Das Colab-Notebook, in dem die Funktion is_car_acc_prob zum Filtern des Datensatzes verwendet wird, ist im GitHub Repository verfügbar.
Bei der Abstimmung der Vorfilterung haben wir Folgendes beobachtet:
- Viele der Autobilder mit hellem Hintergrund wurden als „Strandwagen“ klassifiziert. Wir haben daher entschieden, auch die Klasse „Strandwagen“ in imagenet als eine der
CAR_CLASSESzu berücksichtigen. - Bilder, die die Front eines Autos zeigen, bekommen oft eine hohe Wahrscheinlichkeit der Klasse „Kühlergrill“ („grille“) zugeordnet, d.h. dem Gitter an der Front eines Autos, das zur Kühlung dient. Diese Zuordnung ist korrekt, führt aber dazu, dass die oben gezeigte Prozedur bestimmte Bilder von Autos nicht als Autos betrachtet, da wir „grille“ nicht in die
CAR_CLASSESaufgenommen haben. Dieses Problem führt zu dem Kompromiss, entweder viele Nahaufnahmen von Autokühlergrills im Datensatz zu belassen oder einige Autobilder herauszufiltern. Wir haben uns für den zweiten Ansatz entschieden, da er einen saubereren Datensatz ergibt.
Nach der Vorfilterung der Bilder mit dem vorgeschlagenen Verfahren verbleiben zunächst 53’738 von 64’467 im Datensatz.
Übersicht über die endgültigen Datensätze
Der vorgefilterte Datensatz enthält Bilder von 323 Automodellen. Wir haben uns dazu entschieden, unsere Aufmerksamkeit auf die 300 häufigsten Klassen im Datensatz zu reduzieren. Das ist deshalb sinnvoll, da einige der am wenigsten häufigen Klassen weniger als zehn Repräsentanten haben und somit nicht sinnvoll in ein Trainings-, Validierungs- und Testset aufgeteilt werden können. Reduziert man den Datensatz auf die Bilder der 300 häufigsten Klassen, erhält man einen Datensatz mit 53.536 beschrifteten Bildern. Die Klassenvorkommen sind wie folgt verteilt:

Die Anzahl der Bilder pro Klasse (Automodell) reicht von 24 bis knapp unter 500. Wir können sehen, dass der Datensatz sehr unausgewogen ist. Dies muss beim Training und bei der Auswertung des Modells unbedingt beachtet werden.
Aufbau von Datenpipelines mit tf.data
Selbst nach der Vorfilterung und der Reduktion auf die besten 300 Klassen bleiben immer noch zahlreiche Bilder übrig. Dies stellt ein potenzielles Problem dar, da wir nicht einfach alle Bilder auf einmal in den Speicher unserer GPU laden können. Um dieses Problem zu lösen, werden wir tf.data verwenden.
Mit tf.data und insbesondere der tf.data.Dataset API lassen sich elegante und gleichzeitig sehr effiziente Eingabe-Pipelines erstellen. Die API enthält viele allgemeine Methoden, die zum Laden und Transformieren potenziell großer Datensätze verwendet werden können. Die Methode tf.data.Dataset ist besonders nützlich, wenn Modelle auf GPU(s) trainiert werden. Es ermöglicht das Laden von Daten von der Festplatte, wendet on-the-fly Transformationen an und erstellt Batches, die dann an die GPU gesendet werden. Und das alles geschieht so, dass die GPU nie auf neue Daten warten muss.
Die folgenden Funktionen erstellen eine <code>tf.data.Dataset-Instanz für unseren konkreten Anwendungsfall:
def construct_ds(input_files: list,
batch_size: int,
classes: list,
label_type: str,
input_size: tuple = (212, 320),
prefetch_size: int = 10,
shuffle_size: int = 32,
shuffle: bool = True,
augment: bool = False):
"""
Function to construct a tf.data.Dataset set from list of files
Args:
input_files: list of files
batch_size: number of observations in batch
classes: list with all class labels
input_size: size of images (output size)
prefetch_size: buffer size (number of batches to prefetch)
shuffle_size: shuffle size (size of buffer to shuffle from)
shuffle: boolean specifying whether to shuffle dataset
augment: boolean if image augmentation should be applied
label_type: 'make' or 'model'
Returns:
buffered and prefetched tf.data.Dataset object with (image, label) tuple
"""
# Create tf.data.Dataset from list of files
ds = tf.data.Dataset.from_tensor_slices(input_files)
# Shuffle files
if shuffle:
ds = ds.shuffle(buffer_size=shuffle_size)
# Load image/labels
ds = ds.map(lambda x: parse_file(x, classes=classes, input_size=input_size, label_type=label_type))
# Image augmentation
if augment and tf.random.uniform((), minval=0, maxval=1, dtype=tf.dtypes.float32, seed=None, name=None) < 0.7:
ds = ds.map(image_augment)
# Batch and prefetch data
ds = ds.batch(batch_size=batch_size)
ds = ds.prefetch(buffer_size=prefetch_size)
return dsWir werden nun die verwendeten tf.data-Methoden beschreiben:
from_tensor_slices()ist eine der verfügbaren Methoden für die Erstellung eines Datensatzes. Der erzeugte Datensatz enthält Slices des angegebenen Tensors, in diesem Fall die Dateinamen.- Als nächstes betrachtet die Methode
shuffle()jeweilsbuffer_size-Elemente separat und mischt diese Elemente isoliert vom Rest des Datensatzes. Wenn das Mischen des gesamten Datensatzes erforderlich ist, mussbuffer_sizegrößer sein als die Anzahl der Einträge im Datensatz. Das Mischen wird nur durchgeführt, wennshuffle=Truegesetzt ist. - Mit
map()lassen sich beliebige Funktionen auf den Datensatz anwenden. Wir haben eine Funktionparse_file()erstellt, die im GitHub Repo zu finden ist. Sie ist verantwortlich für das Lesen und die Größenänderung der Bilder, das Ableiten der Beschriftungen aus dem Dateinamen und die Kodierung der Beschriftungen mit einem One-Hot-Encoder. Wenn die Flag „augment“ gesetzt ist, wird das Verfahren zur Datenerweiterung aktiviert. Die Augmentierung wird nur in 70 % der Fälle angewendet, da es von Vorteil ist, das Modell auch auf nicht modifizierten Bildern zu trainieren. Die inimage_augmentverwendeten Augmentierungstechniken sind Flipping, Helligkeits- und Kontrastanpassungen. - Schließlich wird die Methode
batch()verwendet, um den Datensatz in Batches der Größebatch_sizezu gruppieren, und die Methodeprefetch()ermöglicht die Vorbereitung späterer Batches, während der aktuelle Batch verarbeitet wird, und verbessert so die Leistung. Wenn die Methode nach einem Aufruf vonbatch()verwendet wird, werdenprefetch_size-Batches vorab geholt.
Fine Tuning des Modells
Nachdem wir unsere Eingabe-Pipeline definiert haben, wenden wir uns nun dem Trainingsteil des Modells zu. Der Code unten zeigt auf, wie ein Modell basierend auf dem vortrainierten ResNet instanziiert werden kann:
from tensorflow.keras.applications import ResNet50V2
from tensorflow.keras import Model
from tensorflow.keras.layers import Dense, GlobalAveragePooling2D
class TransferModel:
def __init__(self, shape: tuple, classes: list):
"""
Class for transfer learning from ResNet
Args:
shape: Input shape as tuple (height, width, channels)
classes: List of class labels
"""
self.shape = shape
self.classes = classes
self.history = None
self.model = None
# Use pre-trained ResNet model
self.base_model = ResNet50V2(include_top=False,
input_shape=self.shape,
weights='imagenet')
# Allow parameter updates for all layers
self.base_model.trainable = True
# Add a new pooling layer on the original output
add_to_base = self.base_model.output
add_to_base = GlobalAveragePooling2D(data_format='channels_last', name='head_gap')(add_to_base)
# Add new output layer as head
new_output = Dense(len(self.classes), activation='softmax', name='head_pred')(add_to_base)
# Define model
self.model = Model(self.base_model.input, new_output)Ein paar weitere Details zum oben stehenden Code:
- Wir erzeugen zunächst eine Instanz der Klasse
tf.keras.applications.ResNet50V2. Mitinclude_top=Falseweisen wir das vortrainierte Modell an, den ursprünglichen head des Modells (in diesem Fall für die Klassifikation von 1000 Klassen auf ImageNet ausgelegt) wegzulassen. - Mit
base_model.trainable = Truewerden alle Layer trainierbar. - Mit der funktionalen API
tf.kerasstapeln wir dann ein neues Pooling-Layer auf den letzten Faltungsblock des ursprünglichen ResNet-Modells. Dies ist ein notwendiger Zwischenschritt, bevor die Ausgabe an die endgültigen Klassifizierungs-Layer weitergeleitet wird. - Die endgültigen Klassifizierungs-Layer wird dann mit „tf.keras.layers.Dense“ definiert. Wir definieren die Anzahl der Neuronen so, dass sie gleich der Anzahl der gewünschten Klassen ist. Und die Softmax-Aktivierungsfunktion sorgt dafür, dass die Ausgabe eine Pseudowahrscheinlichkeit im Bereich von
(0,1]ist.
Die Vollversion von TransferModel (s. GitHub) enthält auch die Option, das Basismodell durch ein VGG16-Netzwerk zu ersetzen, ein weiteres Standard-CNN für die Bildklassifikation. Außerdem erlaubt es, nur bestimmte Layer freizugeben, d.h. wir können die entsprechenden Parameter trainierbar machen, während wir die anderen festgehalten werden. Standardmässig haben wir hier alle Parameter trainierbar gemacht.
Nachdem wir das Modell definiert haben, müssen wir es für das Training konfigurieren. Dies kann mit der compile()-Methode von tf.keras.Model gemacht werden:
def compile(self, **kwargs):
"""
Compile method
"""
self.model.compile(**kwargs)Wir übergeben dann die folgenden Keyword-Argumente an unsere Methode:
loss = "categorical_crossentropy"für die Mehrklassen-Klassifikation,optimizer = Adam(0.0001)für die Verwendung des Adam-Optimierers austf.keras.optimizersmit einer relativ kleinen Lernrate (mehr zur Lernrate weiter unten), undmetrics = ["categorical_accuracy"]für die Trainings- und Validierungsüberwachung.
Als Nächstes wollen wir uns das Trainingsverfahren ansehen. Dazu definieren wir eine train-Methode für unsere oben vorgestellte TransferModel-Klasse:
from tensorflow.keras.callbacks import EarlyStopping
def train(self,
ds_train: tf.data.Dataset,
epochs: int,
ds_valid: tf.data.Dataset = None,
class_weights: np.array = None):
"""
Trains model in ds_train with for epochs rounds
Args:
ds_train: training data as tf.data.Dataset
epochs: number of epochs to train
ds_valid: optional validation data as tf.data.Dataset
class_weights: optional class weights to treat unbalanced classes
Returns
Training history from self.history
"""
# Define early stopping as callback
early_stopping = EarlyStopping(monitor='val_loss',
min_delta=0,
patience=12,
restore_best_weights=True)
callbacks = [early_stopping]
# Fitting
self.history = self.model.fit(ds_train,
epochs=epochs,
validation_data=ds_valid,
callbacks=callbacks,
class_weight=class_weights)
return self.historyDa unser Modell eine Instanz von tensorflow.keras.Model ist, können wir es mit der Methode fit trainieren. Um Overfitting zu verhindern, wird Early Stopping verwendet, indem es als Callback-Funktion an die fit-Methode übergeben wird. Der patience-Parameter kann eingestellt werden, um festzulegen, wie schnell das Early Stopping angewendet werden soll. Der Parameter steht für die Anzahl der Epochen, nach denen, wenn keine Abnahme des Validierungsverlustes registriert wird, das Training abgebrochen wird. Weiterhin können Klassengewichte an die Methode fit übergeben werden. Klassengewichte erlauben es, unausgewogene Daten zu behandeln, indem den verschiedenen Klassen unterschiedliche Gewichte zugewiesen werden, wodurch die Wirkung von Klassen mit weniger Trainingsbeispielen erhöht werden kann.
Wir können den Trainingsprozess mit einem vortrainierten Modell wie folgt beschreiben: Da die Gewichte im head zufällig initialisiert werden und die Gewichte des Basismodells vortrainiert sind, setzt sich das Training aus dem Training des heads von Grund auf und der Feinabstimmung der Gewichte des vortrainierten Modells zusammen. Es wird generell für Transfer Learning empfohlen, eine kleine Lernrate zu verwenden (z. B. 1e-4), da eine zu große Lernrate die nahezu optimalen vortrainierten Gewichte des Basismodells zerstören kann.
Der Trainingsvorgang kann beschleunigt werden, indem zunächst einige Epochen lang trainiert wird, ohne dass das Basismodell trainierbar ist. Der Zweck dieser ersten Epochen ist es, die Gewichte des heads an das Problem anzupassen. Dies beschleunigt das Training, da wenn nur der head trainiert wird, viel weniger Parameter trainierbar sind und somit für jeden Batch aktualisiert werden. Die resultierenden Modellgewichte können dann als Ausgangspunkt für das Training des gesamten Modells verwendet werden, wobei das Basismodell trainierbar ist. Für das hier betrachtete Autoklassifizierungsproblem führte die Anwendung dieses zweistufigen Trainings zu keiner nennenswerten Leistungsverbesserung.
Evaluation/Vorhersage der Modell Performance
Bei der Verwendung der API tf.data.Dataset muss man auf die Art der verwendeten Methoden achten. Die folgende Methode in unserer Klasse TransferModel kann als Vorhersagemethode verwendet werden.
def predict(self, ds_new: tf.data.Dataset, proba: bool = True):
"""
Predict class probs or labels on ds_new
Labels are obtained by taking the most likely class given the predicted probs
Args:
ds_new: New data as tf.data.Dataset
proba: Boolean if probabilities should be returned
Returns:
class labels or probabilities
"""
p = self.model.predict(ds_new)
if proba:
return p
else:
return [np.argmax(x) for x in p]Es ist wichtig, dass der Datensatz ds_new nicht gemischt wird, sonst stimmen die erhaltenen Vorhersagen nicht mit den erhaltenen Bildern überein, wenn ein zweites Mal über den Datensatz iteriert wird. Dies ist der Fall, da die Flag reshuffle_each_iteration in der Implementierung der Methode shuffle standardmäßig auf True gesetzt ist. Ein weiterer Effekt des Shufflens ist, dass mehrere Aufrufe der Methode take nicht die gleichen Daten zurückgeben. Dies ist wichtig, wenn z. B. Vorhersagen für nur eine Charge überprüft werden sollen. Ein einfaches Beispiel, an dem dies zu sehen ist, ist:
# Use construct_ds method from above to create a shuffled dataset
ds = construct_ds(..., shuffle=True)
# Take 1 batch (e.g. 32 images) of dataset: This returns a new dataset
ds_batch = ds.take(1)
# Predict labels for one batch
predictions = model.predict(ds_batch)
# Predict labels again: The result will not be the same as predictions above due to shuffling
predictions_2 = model.predict(ds_batch)Eine Funktion zum Plotten von Bildern, die mit den entsprechenden Vorhersagen beschriftet sind, könnte wie folgt aussehen:
def show_batch_with_pred(model, ds, classes, rescale=True, size=(10, 10), title=None):
for image, label in ds.take(1):
image_array = image.numpy()
label_array = label.numpy()
batch_size = image_array.shape[0]
pred = model.predict(image, proba=False)
for idx in range(batch_size):
label = classes[np.argmax(label_array[idx])]
ax = plt.subplot(np.ceil(batch_size / 4), 4, idx + 1)
if rescale:
plt.imshow(image_array[idx] / 255)
else:
plt.imshow(image_array[idx])
plt.title("label: " + label + "n"
+ "prediction: " + classes[pred[idx]], fontsize=10)
plt.axis('off')Die Methode show_batch_with_pred funktioniert auch für gemischte Datensätze, da image und label demselben Aufruf der Methode take entsprechen.
Die Auswertung der Model-Performance kann mit der Methode evaluate von keras.Model durchgeführt werden.
Wie akkurat ist unser finales Modell?
Das Modell erreicht eine kategoriale Genauigkeit von etwas über 70 % für die Vorhersage des Automodells für Bilder aus 300 Modellklassen. Um die Vorhersagen des Modells besser zu verstehen, ist es hilfreich, die Konfusionsmatrix zu betrachten. Unten ist die Heatmap der Vorhersagen des Modells für den Validierungsdatensatz abgebildet.
Wir haben die Heatmap auf Einträge der Konfusionsmatrix in [0, 5] beschränkt, da das Zulassen einer weiteren Spanne keine Region außerhalb der Diagonalen signifikant hervorgehoben hat. Wie in der Heatmap zu sehen ist, wird eine Klasse den Beispielen fast aller Klassen zugeordnet. Das ist an der dunkelroten vertikalen Linie zwei Drittel rechts in der Abbildung oben zu erkennen.
Abgesehen von der zuvor erwähnten Klasse gibt es keine offensichtlichen Verzerrungen in den Vorhersagen. Wir möchten an dieser Stelle betonen, dass die Accuracy im Allgemeinen nicht ausreicht, um die Leistung eines Modells zufriedenstellend zu beurteilen, insbesondere im Fall unausgewogener Klassen.
Fazit und nächste Schritte
In diesem Blog-Beitrag haben wir Transfer Learning mit dem ResNet50V2 angewendet, um das Fahrzeugmodell anhand von Bildern von Autos zu klassifizieren. Unser Modell erreicht 70% kategoriale Genauigkeit über 300 Klassen.
Wir haben festgestellt, dass das Trainieren des gesamten Basismodells und die Verwendung einer kleinen Lernrate die besten Ergebnisse erzielen. Ein cooles Auto-Klassifikationsmodell entwickelt zu haben ist großartig, aber wie können wir unser Modell in einer produktiven Umgebung einsetzen? Natürlich könnten wir unsere eigene Modell-API mit Flask oder FastAPI bauen… Aber gibt es vielleicht sogar einen einfacheren, standardisierten Weg?
Im zweiten Beitrag unserer Blog-Serie, „Deployment von TensorFlow-Modellen in Docker mit TensorFlow Serving“ zeigen wir Euch, wie dieses Modell mit TensorFlow Serving bereitgestellt werden kann.
Management Summary
OCR (Optical Character Recognition) ist eine große Herausforderung für viele Unternehmen. Am OCR-Markt tummeln sich diverse Open Source sowie kommerzielle Anbieter. Ein bekanntes Open Source Tool für OCR ist Tesseract, das mittlerweile von Google bereitgestellt wird. Tesseract ist aktuell in der Version 4 verfügbar, die die OCR Extraktion mittels rekurrenten neuronalen Netzen durchführt. Die OCR Performance von Tesseract ist nach wie vor jedoch volatil und hängt von verschiedenen Faktoren ab. Eine besondere Herausforderung ist die Anwendung von Tesseract auf Dokumente, die aus verschiedenen Strukturen aufgebaut sind, z.B. Texten, Tabellen und Bildern. Eine solche Dokumentenart stellen bspw. Rechnungen dar, die OCR Tools aller Anbieter nach wie vor besondere Herausforderungen stellen.
In diesem Beitrag wird demonstriert, wie ein Finetuning der Tesseract-OCR (Optical Character Recognition) Engine auf einer kleinen Stichprobe von Daten bereits eine erhebliche Verbesserung der OCR-Leistung auf Rechnungsdokumenten bewirken kann. Dabei ist der dargestellte Prozess nicht ausschließlich auf Rechnungen anwendbar sondern auf beliebige Dokumentenarten.
Es wird ein Anwendungsfall definiert, der auf eine korrekte Extraktion des gesamten Textes (Wörter und Zahlen) aus einem fiktiven, aber realistischen deutschen Rechnungsdokument abzielt. Es wird hierbei angenommen, dass die extrahierten Informationen für nachgelagerte Buchhaltungszwecke bestimmt sind. Daher wird eine korrekte Extraktion der Zahlen sowie des Euro-Zeichens als kritisch angesehen.
Die OCR-Leistung von zwei Tesseract-Modellen für die deutsche Sprache wird verglichen: das Standardmodell (nicht getuned) und eine finegetunete Variante. Das Standardmodell wird aus dem Tesseract OCR GitHub Repository bezogen. Das feinabgestimmte Modell wird mit denen in diesem Artikel beschriebenen Schritten entwickelt. Eine zweite deutsche Rechnung ähnlich der ersten wird für die Feinabstimmung verwendet. Sowohl das Standardmodell als auch das getunte Modell werden auf der gleichen Out-of-Sample Rechnung bewertet, um einen fairen Vergleich zu gewährleisten.
Die OCR-Leistung des Tesseract Standardmodells ist bei Zahlen vergleichsweise schlecht. Dies gilt insbesondere für Zahlen, die den Zahlen 1 und 7 ähnlich sind. Das Euro-Symbol wird in 50% der Fälle falsch erkannt, sodass das Ergebnis für eine etwaig nachgelagerte Buchhaltungsanwendung ungeeignet ist.
Das getunte Modell zeigt eine ähnliche OCR-Leistung für deutsche Wörter. Die OCR-Leistung bei Zahlen verbessert sich jedoch deutlich. Alle Zahlen und jedes Euro-Symbol werden korrekt extrahiert. Es zeigt sich, dass eine Feinabstimmung mit minimalem Aufwand und einer geringen Menge an Schulungsdaten eine große Verbesserung der Erkennungsleistung erzielen kann. Dadurch wird Tesseract OCR mit seiner Open-Source-Lizenzierung zu einer attraktiven Lösung im Vergleich zu proprietärer OCR-Software. Weiterhin werden abschließende Empfehlungen für das Finetuning von Tesseract LSTM-Modellen dargestellt, für den Fall, dass mehr Trainingsdaten vorliegen.
Einführung
Tesseract 4 mit seiner LSTM-Engine funktioniert out-of-the-box für einfache Texte bereits recht gut. Es gibt jedoch Szenarien, für die das Standardmodell schlecht abschneidet. Beispiele hierfür sind exotische Schriftarten, Bilder mit Hintergründen oder Text in Tabellen. Glücklicherweise bietet Tesseract eine Möglichkeit zum Finetuning der LSTM-Engine, um die OCR-Leistung für speziellere Anwendungsfälle zu verbessern.
Warum OCR für Rechnungen eine Herausforderung ist
Auch wenn OCR in Teilbereichen als ein gelöstes Problem gilt, stellt die fehlerfreie Extraktion eines großen Textkorpus nach wie vor eine Herausforderung dar. Dies gilt insbesondere für OCR auf Dokumenten, die eine hohe strukturelle Varianz aufweisen, wie bspw. Rechnungsdokumente. Diese bestehen häufig aus unterschiedlichsten Elementen, die OCR-Engine von Tesseract for Herausforderungen stellen:
1. Farbige Hintergründe und Tabellenstrukturen stellen eine Herausforderung für die Seitensegmentierung dar.
2. Rechnungen enthalten normalerweise seltene Zeichen wie das EUR- oder USD-Zeichen
3. Zahlen können nicht mit einem Sprachwörterbuch überprüft werden.
Darüber hinaus ist die Fehlermarge gering: Häufig ist eine exakte Extraktion der numerischen Daten für nachfolgenden Prozessschritte von größter Bedeutung.
Problem (1) lässt sich in der Regel dadurch lösen, dass man eine der 14 von Tesseract bereitgestellten Segmentierungsmodus auswählt. Die beiden letztgenannten Probleme lassen sich häufig durch ein Finetuning der LSTM-Engine auf Basis von Beispielen ähnlicher Dokumente lösen.
Use Case Zielsetzung und Daten
Zwei ähnliche Beispielrechnungen werden in dem Artikel näher betrachtet. Die in Abbildung 1 gezeigte Rechnung wird zur Bewertung der OCR-Leistung sowohl für das Standard- als auch des feingetunte Tesseract-Modell verwendet. Besondere Aufmerksamkeit wird der korrekten Extraktion von Zahlen gewidmet. Die in Abbildung 2 gezeigte, zweite Rechnung wird zum Finetuning des LSTM Modells verwendet.
Die meisten Rechnungsdokumente sind in einer sehr gut lesbaren Schriftart wie „Arial“ geschrieben. Um die Vorteile des Tunings zu veranschaulichen, wird das anfängliche OCR-Problem durch die Berücksichtigung von Rechnungen, die in der Schriftart „Impact“ geschrieben sind, erschwert. „Impact“ ist eine Schriftart, die sich deutlich von normalen serifenlosen Schriften unterscheidet, und zu einer höheren Fehlerkennung für Tesseract führt.
Es wird im Folgenden gezeigt, dass Tesseract nach der Feinabstimmung auf Basis einer sehr kleinen Datenmenge trotz dieser schwierigen Schriftart sehr zufriedenstellende Ergebnisse liefert.
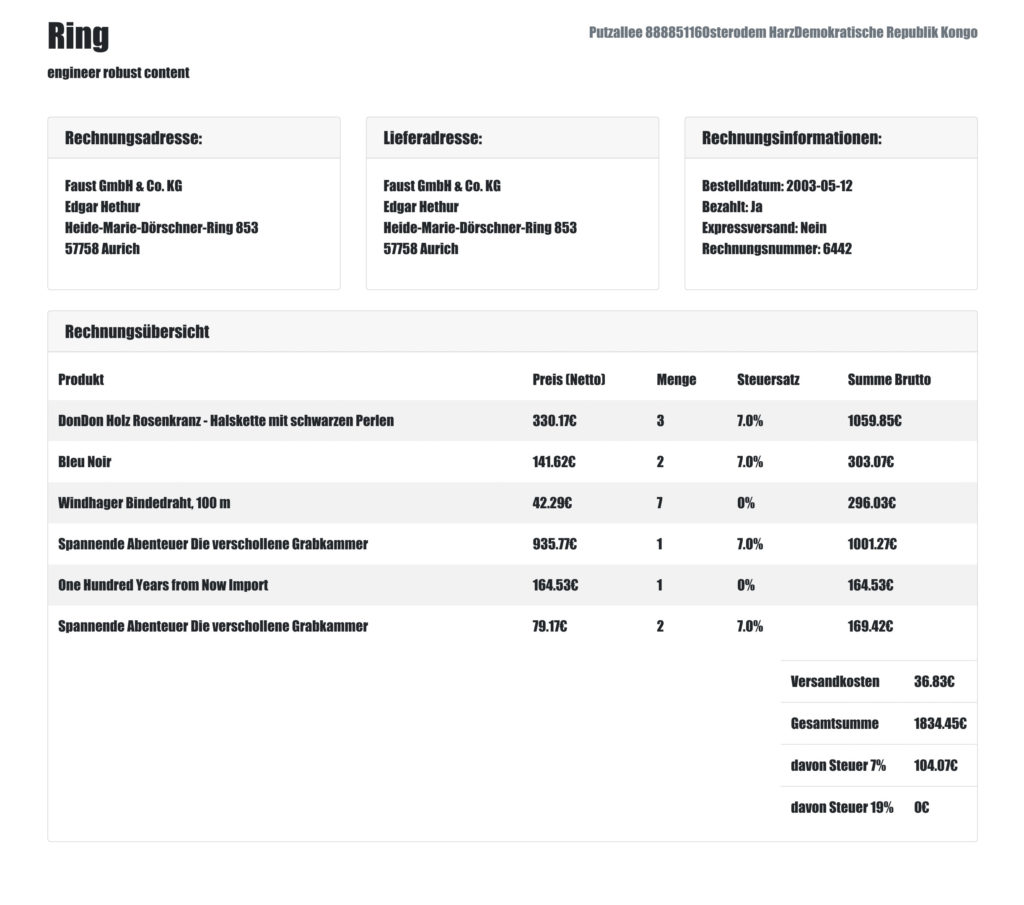
Verwendung des Tesseract 4.1 Docker Containers
Die Einrichtung zum Finetuning der Tesseract-LSTM-Engine funktioniert derzeit nur unter Linux und kann etwas knifflig sein. Daher wird zusammen mit diesem Artikel ein Docker-Container mit vorinstalliertem Tesseract 4.1 sowie mit den kompilierten Trainings-Tools und Skripten bereitgestellt.
Mit folgenden Befehlen ziehen Sie das Container Image von Docker Hub auf Ihren Rechner und rufen die Shell im Container auf. Nun können Sie die Schritte aus diesem Blogpost replizieren.
docker pull statworx/blog-tesseract
docker run -it --name tesseract_container statworx/blog-tesseract /bin/bash Allgemeine Verbesserungen der OCR Performance
Es gibt drei Möglichkeiten, wie die OCR-Leistung von Tesseract verbessert werden kann, noch bevor ein Finetuning der LSTM-Engine vorgenommen wird.
1. Preprocessing der Bilder
Gescannte Dokumente können eine schiefe Ausrichtung haben, wenn sie auf dem Scanner nicht richtig platziert wurden. Gedrehte Bilder sollten entzerrt werden, um die Liniensegmentierungsleistung von Tesseract zu optimieren.
Darüber hinaus kann beim Scannen ein Bildrauschen entstehen, das durch einen Rauschunterdrückungsalgorithmus entfernt werden sollte. Beachten Sie, dass Tesseract standardmäßig eine Schwellenwertbildung unter Verwendung des Otsu-Algorithmus durchführt, um Graustufenbilder in schwarze und weiße Pixel zu binärisieren.
Eine detaillierte Behandlung der Bildvorverarbeitung würde den Rahmen dieses Artikels sprengen und ist nicht notwendig, um für den gegebenen Anwendungsfall zufriedenstellende Ergebnisse zu erzielen. Die Tesseract-Dokumentation bietet einen praktischen Überblick.
2. Seitensegmentierung
Während der Seitensegmentierung versucht Tesseract, rechteckige Textbereiche zu identifizieren. Nur diese Bereiche werden im nächsten Schritt für die OCR ausgewählt. Es ist daher wichtig, alle Regionen mit Text zu erfassen, damit keine Informationen verloren gehen.
Tesseract ermöglicht die Auswahl aus 14 verschiedenen Seitensegmentierungsmethoden, die mit dem folgenden Befehl angezeigt werden können:
tesseract --help-psmDie Standard-Segmentierungsmethode erwartet eine Bild ähnlich zu einer Buchseite. Dieser Modus kann jedoch aufgrund der zusätzlichen tabellarischen Strukturen in Rechnungsdokumenten nicht alle Textbereiche korrekt identifizieren. Eine bessere Segmentierungsmethode ist durch Option 4 gegeben: „Assume a single column of text of variable sizes“.
Um die Bedeutung einer geeigneten Seitensegmentierungsmethode zu veranschaulichen, betrachten wir das Ergebnis der Verwendung der Standardmethode „Fully automatic page segmentation, but no OSD “ in Abbildung 3:

Abbildung 3: Die Standard-Segmentierungsmethode kann nicht alle Textbereiche erkennen
Beachten Sie, dass die Texte „Rechnungsinformationen:“, „Pos.“ und „Produkt“ nicht segmentiert wurden. In Abbildung 4 führt eine geeignetere Methode zu einer perfekten Segmentierung der Seite.
3. Verwendung von Dictionaries, Wortlisten und Mustern für den Text
Die von Tesseract verwendeten LSTM-Modelle wurden auf Basis von großen Textmengen in einer bestimmten Sprache trainiert. Dieser Befehl zeigt die Sprachen an, die derzeit für Tesseract verfügbar sind:
tesseract --list-langs Weitere Sprachmodelle sind verfügbar, indem die entsprechenden language.tessdata heruntergelden und in den Ordner tessdata der lokalen Tesseract-Installation abgelegt werden. Das Tesseract-Repository auf GitHub stellt drei Varianten von Sprachmodellen zur Verfügung: normal, fast und best. Nur die schnelle sowie die beste Variante sind für ein Finetuning verwendbar. Wie der Name schon sagt, handelt es sich dabei um die schnellsten bzw. genauesten Varianten von Modellen. Weitere Modelle wurden ebenfalls für spezielle Anwendungsfälle wie die ausschließliche Erkennung von Ziffern und Interpunktion trainiert und sind in den Referenzen aufgeführt.
Da die Sprache der Rechnungen in diesem Anwendungsfall Deutsch ist, wird das zu diesem Artikel gehörende Docker-Image mit dem deu.tessdata-Modell geliefert.
Für eine bestimmte Sprache kann die Wortliste von Tesseract weiter ausgebaut oder auf bestimmte Wörter oder sogar Zeichen beschränkt werden. Dieses Thema liegt außerhalb des Rahmens dieses Artikels, da es nicht notwendig ist, um für den vorliegenden Anwendungsfall zufriedenstellende Ergebnisse zu erzielen.
Setup des Finetuning-Prozesses
Für das Finetuning müssen drei Dateitypen erstellt werden:
1. tiff-Dateien
Tagged Image File Format oder TIFF ist ein unkomprimiertes Bilddateiformat (im Gegensatz zu JPG oder PNG, die komprimierte Dateiformate sind). TIFF-Dateien können mit einem Konvertierungswerkzeug aus PNG- oder JPG-Formaten gewonnen werden. Obwohl Tesseract mit PNG- und JPG-Bildern arbeiten kann, wird das TIFF-Format empfohlen.
2. Box-Dateien
Zum Trainieren des LSTM-Modells verwendet Tesseract so genannte Box-Dateien mit der Erweiterung „.box“. Eine Box-Datei enthält den erkannten Text zusammen mit den Koordinaten der Bounding Box, in der sich der Text befindet. Box-Dateien enthalten sechs Spalten, die korrespondieren zu Symbol, Links, Unten, Rechts, Oben und Seite:
P 157 2566 1465 2609 0
r 157 2566 1465 2609 0
o 157 2566 1465 2609 0
d 157 2566 1465 2609 0
u 157 2566 1465 2609 0
k 157 2566 1465 2609 0
t 157 2566 1465 2609 0
157 2566 1465 2609 0
P 157 2566 1465 2609 0
r 157 2566 1465 2609 0
e 157 2566 1465 2609 0
i 157 2566 1465 2609 0
s 157 2566 1465 2609 0
157 2566 1465 2609 0
( 157 2566 1465 2609 0
N 157 2566 1465 2609 0
e 157 2566 1465 2609 0
t 157 2566 1465 2609 0
t 157 2566 1465 2609 0
o 157 2566 1465 2609 0
) 157 2566 1465 2609 0
157 2566 1465 2609 0
Jedes Zeichen befindet sich auf einer separaten Zeile in der Box-Datei. Das LSTM-Modell akzeptiert entweder die Koordinaten einzelner Zeichen oder einer ganzen Textzeile. In der obigen Beispiel-Box-Datei befindet sich der Text „Produkt Preis (Netto)“ optisch auf der gleichen Zeile im Dokument. Alle Zeichen haben die gleichen Koordinaten, nämlich die Koordinaten des Begrenzungsrahmens um diese Textzeile herum. Die Verwendung von Koordinaten auf Zeilenebene ist wesentlich einfacher und wird standardmäßig bereitgestellt, wenn die Box-Datei mit dem folgenden Befehl erzeugt wird:
cd /home/fine_tune/train
tesseract train_invoice.tiff train_invoice --psm 4 -l best/deu lstmboxDas erste Argument ist die zu extrahierende Bilddatei, das zweite Argument stellt den Dateinamen der Box-Datei dar. Der Sprachparameter -l weist Tesseract an, das deutsche Modell für die OCR zu verwenden. Der Parameter –psm weist Tesseract an, das vierte Seitensegmentierungsverfahren zu verwenden.
Nahezu unvermeidlich ist, dass die generierten OCR-Box-Dateien Fehler in der Symbolspalte enthalten. Jedes Symbol in der Box-Datei des Trainings muss daher von Hand überprüft werden. Dies ist ein mühsamer Prozess, da die Box-Datei der Demo-Rechnung fast tausend Zeilen enthält (eine für jedes Zeichen in der Rechnung). Um die Korrektur zu vereinfachen, stellt der Docker-Container ein Python-Skript zur Verfügung, das die Bounding-Boxes zusammen mit dem OCR-Text auf dem Originalrechnungsbild zeichnet, um einen Vergleich zwischen der Box Datei und dem Dokument zu erleichtern. Das Ergebnis ist in Abbildung 4 dargestellt. Der Docker-Container enthält bereits die korrigierten Box-Dateien, die durch den Suffix „_correct“ gekennzeichnet sind.

Abbildung 4 – Extrahierter Text bei Anwendung des Tesseract Modells „deu“
3. lstmf Dateien
Während des Finetunings extrahiert Tesseract den Text aus der Tiff-Datei und überprüft die Vorhersage anhand der Koordinaten sowie des Symbols in der Box-Datei. Tesseract verwendet dabei nicht direkt die Tiff- und Box-Datei, sondern erwartet eine sog. lstmf-Datei, die aus den beiden vorherigen Dateien erstellt wurde. Hierbei ist zu beachten, dass zur Erstellung der lstmf-Datei die Tiff- und Box-Datei denselben Namen haben müssen, z.B. train_invoice.tiff und train_invoice.box.
Der folgende Befehl erzeugt eine lstmf-Datei für die Zugrechnung:
cd /home/fine_tune/train
tesseract train_invoice.tiff train_invoice lstm.train Alle lstmf-Dateien, die für das Training relevant sind, müssen durch ihren relativen Pfad in einer Textdatei namens deu.training_files.txt angegeben werden. In diesem Anwendungsfall wird nur eine lstmf-Datei für das Training verwendet, so dass die Datei deu.training_files.txt nur eine Zeile enthält, nämlich: eval/train_invoice_correct.lstmf.
Es wird empfohlen, auch eine lstfm-Datei für die Evaluierungs-Rechnung zu erstellen. Auf diese Weise kann die Performance des Modells während dem Trainingsvorgang bewertet werden:
cd /home/fine_tune/eval
tesseract eval_invoice_correct.tiff eval_invoice_correct lstm.trainEvaluierung des Standard-LSTM-Modells
OCR-Vorhersagen aus dem deutschen Standardmodell „deu“ werden als Benchmark verwendet. Einen genauen Überblick über die OCR-Leistung des deutschen Standardmodells erhält man, indem man eine Box-Datei für die Evaluierungs-Rechnung erzeugt und den OCR-Text mit dem bereits erwähnten Python-Skript visualisiert. Dieses Skript, das die Datei „eval_invoice_ocr deu.tiff“ erzeugt, befindet sich im mitgelieferten Container unter „/home/fine_tune/src/draw_box_file_data.py“. Das Skript erwartet als Argument den Pfad zu einer Tiff-Datei, die entsprechende Box-Datei sowie einen Namen für die Ausgabe-Tiff-Datei. Der durch das deutsche Standardmodell extrahierte OCR-Text wird als eval/eval_invoice_ocr_deu.tiff gespeichert und ist in Abbildung 1 dargestellt.
Auf den ersten Blick sieht der durch OCR extrahierte Text gut aus. Das Modell extrahiert deutsche Zeichen wie ä, ö ü und ß korrekt. Tatsächlich gibt es nur drei Fälle, in denen Wörter Fehler enthalten:
| OCR | Truth |
| Jessel GmbH 8 Co | Jessel GmbH & Co |
| 11 Glasbehälter | 1l Glasbehälter |
| Zeki64@hloch.com | Zeki64@bloch.com |
Das Modell schneidet bei gebräuchlichen deutschen Wörtern bereits gut ab, hat aber Schwierigkeiten mit singulären Symbolen wie „&“ und „l“ sowie Wörtern wie „bloch“, die nicht in der Wortliste des Modells enthalten sind.
Preise und Zahlen sind für das Modell eine viel größere Herausforderung. Hierbei treten deutlich häufiger Fehler bei der Extraktion auf:
| OCR | Truth |
| 159,16 | 159,1€ |
| 1% | 7% |
| 1305.816 | 1305.81€ |
| 227.66 | 227.6€ |
| 341.51 | 347.57€ |
| 1115.16 | 1115.7€ |
| 242.86 | 242.8€ |
| 1456.86 | 1456.8€ |
| 51.46 | 54.1€ |
| 1954.719€ | 1954.79€ |
Das deutsche Standardmodell extrahiert das Euro-Symbol € in 9 von 18 Fällen nicht korrekt. Dies entspricht einer Fehlerquote von 50%.
Finetuning des Standard-LSTM-Modells
Das Standard-LSTM-Modell wird nun auf der in Abbildung 2 gezeigten Rechnung finegetuned. Anschließend wird die OCR-Leistung anhand der in Abbildung 1 gezeigten Evaluierungs-Rechnung bewertet, die auch zuvor für das Benchmarking des deutschen Standardmodells verwendet wurde.
Zum Finetuning des LSTM-Modells muss dieses zunächst aus der Datei deu.traineddata extrahiert werden. Mit dem folgenden Befehl wird das LSTM-Modell aus dem deutschen Standardmodell in das Verzeichnis lstm_model extrahiert:
cd /home/fine_tune
combine_tessdata -e tesseract/tessdata/best/deu.traineddata lstm_model/deu.lstmAnschließend werden alle notwendigen Dateien für das Finetuning zusammengestellt. Die Dateien sind ebenfalls im Docker-Container vorhanden:
- Die Trainings-Dateien train_invoice_correct.lstmf und deu.training_files.txt im Verzeichnis train.
- Die Evaluierungs-Dateien eval_invoice_correct.lstmf und deu.training_files.txt im eval-Verzeichnis.
- Das extrahierte LSTM-Modell deu.lstm im Verzeichnis lstm_model.
Der Docker-Container enthält das Skript src/fine_tune.sh, das den Prozess des Finetunings startet. Sein Inhalt ist:
/usr/local/bin/lstmtraining \
--model_output output/fine_tuned \
--continue_from lstm_model/deu.lstm \
--traineddata tesseract/tessdata/best/deu.traineddata \
--train_listfile train/deu.training_files.txt \
--eval_listfile eval/deu.training_files.txt \
--max_iterations 400
Mit diesem Befehl wird das extrahierte Modell deu.lstm in der in train/deu.training_files.txt angegebenen Datei train_invoice.lstmf getuned. Das Finetuning des LSTM-Modells erfordert sprachspezifische Informationen, die im Ordner deu.tessdata enthalten sind. Die Datei eval_invoice.lstmf, die in eval/deu.training_files.txt angegeben ist, wird zur Berechnung der OCR-Performance während des Trainings verwendet. Das Finetuning wird nach 400 Iterationen beendet. Die gesamte Trainingsdauer dauert weniger als zwei Minuten.
Der folgende Befehl führt das Skript aus und protokolliert die Ausgabe in einer Datei:
cd /home/fine_tune
sh src/fine_tune.sh > output/fine_tune.log 2>&1Der Inhalt der Protokolldatei nach dem Training ist unten dargestellt:
src/fine_tune.log
Loaded file lstm_model/deu.lstm, unpacking...
Warning: LSTMTrainer deserialized an LSTMRecognizer!
Continuing from lstm_model/deu.lstm
Loaded 20/20 lines (1-20) of document train/train_invoice_correct.lstmf
Loaded 24/24 lines (1-24) of document eval/eval_invoice_correct.lstmf
2 Percent improvement time=69, best error was 100 @ 0
At iteration 69/100/100, Mean rms=1.249%, delta=2.886%, char train=8.17%, word train=22.249%, skip ratio=0%, New best char error = 8.17 Transitioned to stage 1 wrote best model:output/deu_fine_tuned8.17_69.checkpoint wrote checkpoint.
-----
2 Percent improvement time=62, best error was 8.17 @ 69
At iteration 131/200/200, Mean rms=1.008%, delta=2.033%, char train=5.887%, word train=20.832%, skip ratio=0%, New best char error = 5.887 wrote best model:output/deu_fine_tuned5.887_131.checkpoint wrote checkpoint.
-----
2 Percent improvement time=112, best error was 8.17 @ 69
At iteration 181/300/300, Mean rms=0.88%, delta=1.599%, char train=4.647%, word train=17.388%, skip ratio=0%, New best char error = 4.647 wrote best model:output/deu_fine_tuned4.647_181.checkpoint wrote checkpoint.
-----
2 Percent improvement time=159, best error was 8.17 @ 69
At iteration 228/400/400, Mean rms=0.822%, delta=1.416%, char train=4.144%, word train=16.126%, skip ratio=0%, New best char error = 4.144 wrote best model:output/deu_fine_tuned4.144_228.checkpoint wrote checkpoint.
-----
Finished! Error rate = 4.144Während des Trainings speichert Tesseract nach jeder Iteration einen sog. Model Checkpoint. Die Leistung des Modells an diesem Kontrollpunkt wird anhand der Evaluierungs-Daten getestet und mit dem aktuell besten Ergebnis verglichen. Wenn sich das Ergebnis verbessert, d.h. der OCR-Fehler abnimmt, wird eine beschriftete Kopie des Checkpoints gespeichert. Die erste Nummer des Dateinamens für den Kontrollpunkt steht für den Zeichenfehler und die zweite Nummer für die Trainingsiteration.
Der letzte Schritt ist die neue Zusammenstellung des finegetunten LSTM-Modells, so dass man wieder ein „traineddata“ Modell erhält. Unter der Annahme, dass der Kontrollpunkt bei der 181. Iteration selektiert wurde, wird mit dem folgenden Befehl ein ausgewählter Kontrollpunkt „deu_fine_tuned4.647_181.checkpoint“ in ein voll funktionsfähiges Tesseract-Modell „deu_fine_tuned.traineddata“ umgewandelt:
cd /home/fine_tune
/usr/local/bin/lstmtraining \
--stop_training \
--continue_from output/deu_fine_tuned4.647_181.checkpoint \
--traineddata tesseract/tessdata/best/deu.traineddata \
--model_output tesseract/tessdata/best/deu_fine_tuned.traineddata Dieses Modell muss in die Testdaten der lokalen Tesseract-Installation kopiert werden, um es Tesseract zur Verfügung zu stellen. Dies ist im Docker-Container bereits geschehen.
Vergewissern Sie sich, dass das feinabgestimmte Modell in Tesseract verfügbar ist:
tesseract --list-langsEvaluierung des finegetunten LSTM-Modells
Das finegetunte Modell wird analog zum Standardmodell evaluiert: Es wird eine Box-Datei der Auswertungs-Rechnung erstellt, und der OCR-Text wird mit Hilfe des Python-Skripts auf dem Bild der Auswertungsrechnung angezeigt.
Der Befehl zur Erzeugung der Box-Dateien muss so modifiziert werden, dass das fein abgestimmte Modell „deu_fine_tuned“ anstelle des Standardmodells „deu“ verwendet wird:
cd /home/fine_tune/eval
tesseract eval_invoice.tiff eval_invoice --psm 4 -l best/deu_fine_tuned lstmbox Der durch das fein abgestimmte Modell extrahierte OCR-Text ist in Abbildung 5 unten dargestellt.

Wie beim deutschen Standardmodell bleibt die Leistung bei den deutschen Wörtern gut, aber nicht perfekt. Um die Leistung bei seltenen Wörtern zu verbessern, könnte die Wortliste des Modells um weitere Worte erweitert werden.
| OCR | Truth |
| Jessel GmbH 8 Co | Jessel GmbH & Co |
| 1! Glasbehälte | 1l Glasbehälter |
| Zeki64@hloch.com | Zeki64@bloch.com |
Wichtiger ist, dass sich die OCR-Leistung bei Zahlen deutlich verbessert hat: Das verfeinerte Modell extrahierte alle Zahlen und jedes Vorkommen des €-Zeichens korrekt.
| OCR | Truth |
| 159,1€ | 159,1€ |
| 7% | 7% |
| 1305.81€ | 1305.81€ |
| 227.6€ | 227.6€ |
| 347.57€ | 347.57€ |
| 1115.7€ | 1115.7€ |
| 242.8€ | 242.8€ |
| 1456.8€ | 1456.8€ |
| 54.1€ | 54.1€ |
| 1954.79€ | 1954.79€ |
Fazit und Ausblick
In diesem Artikel wurde gezeigt, dass die OCR-Leistung von Tesseract durch Finetuning erheblich verbessert werden kann. Insbesondere bei Nicht-Standard-Anwendungsfällen, wie der Text-Extraktion von Rechnungsdokumenten, kann so die OCR-Leistung signifikant verbessert werden. Neben der Open Source Lizensierung macht die Möglichkeit, die LSTM-Engine von Tesseract mittels Finetunings für spezifische Anwendungsfälle zu tunen, das Framework zu einem attraktiven Tool, auch für anspruchsvollere OCR-Einsatzszenarien.
Zur weiteren Verbesserung des Ergebnisses kann es sinnvoll sein, das Modell für weitere Iterationen zu tunen. In diesem Anwendungsfall wurde die Anzahl der Iterationen absichtlich begrenzt, da nur ein Dokument zum Finetuning verwendet wurde. Mehr Iterationen erhöhen potenziell das Risiko einer Überanpassung des LSTM-Modells auf bestimmten Symbolen, was wiederum die Fehlerquote bei anderen Symbolen erhöht. In der Praxis ist es wünschenswert, die Anzahl der Iterationen unter der Voraussetzung zu erhöhen, dass ausreichend Trainingsdaten zur Verfügung stehen. Die endgültige OCR-Leistung sollte immer auf Basis eines weiteren, jedoch repräsentativen Datensatz von Dokumenten überprüft werden.
Referenzen
- Tesseract training: https://tesseract-ocr.github.io/tessdoc/TrainingTesseract-4.00.html
- Image processing overview: https://tesseract-ocr.github.io/tessdoc/ImproveQuality#image-processing
- Otsu thresholding: https://opencv-python-tutroals.readthedocs.io/en/latest/py_tutorials/py_imgproc/py_thresholding/py_thresholding.html
- Tesseract digits comma model: https://github.com/Shreeshrii/tessdata_shreetest
Did you know, that you can transform plain old static ggplot graphs to animated ones? Well, you can with the help of the package gganimate by RStudio’s Thomas Lin Pedersen and David Robinson and the results are amazing! My STATWORX colleagues and I are very impressed how effortless all kinds of geoms are transformed to suuuper smooth animations. That’s why in this post I will provide a short overview of some of the wonderful functionalities of gganimate, I hope you’ll enjoy them as much as we do!
Since Valentine’s Day is just around the corner, we’re going to explore the Speed Dating Experiment dataset compiled by Columbia Business School professors Ray Fisman and Sheena Iyengar. Hopefully, we’ll learn about gganimate as well as how to find our Valentine. If you like, you can download the data from Kaggle.
Defining the basic animation: transition_*
How are static plots put into motion? Essentially, gganimate creates data subsets, which are plotted individually and constitute the substantial frames, which, when played consecutively, create the basic animation. The results of gganimate are so seamless because gganimate takes care of the so-called tweening for us by calculating data points for transition frames displayed in-between frames with actual input data.
The transition_* functions define how the data subsets are derived and thus define the general character of any animation. In this blogpost we’re going to explore three types of transitions: transition_states(), transition_reveal() and transition_filter(). But let’s start at the beginning.
We’ll start with transition_states(). Here the data is split into subsets according to the categories of the variable provided to the states argument. If several rows of a dataset pertain to the same unit of observation and should be identifiable as such, a grouping variable defining the observation units needs to be supplied. Alternatively, an identifier can be mapped to any other aesthetic.
Please note, to ensure the readability of this post, all text concerning the interpretation of the speed dating data is written in italics. If you’re not interested in that part you simply can skip those paragraphs. For the data prep, I’d like to refer you to my GitHub.
First, we’re going to explore what the participants of the Speed Dating Experiment look for in a partner. Participants were asked to rate the importance of attributes in a potential date by allocating a budget of 100 points to several characteristics, with higher values denoting a higher importance. The participants were asked to rate the attributes according to their own views. Further, the participants were asked to rate the same attributes according to the presumed wishes of their same-sex peers, meaning they allocated the points in the way they supposed their average same-sex peer would do.
We’re going to plot all of these ratings (x-axis) for all attributes (y-axis). Since we want to compare the individual wishes to the individually presumed wishes of peers, we’re going to transition between both sets of ratings. Color always indicates the personal wishes of a participant. A given bubble indicates the rating of one specific participant for a given attribute, switching between one’s own wishes and the wishes assumed for peers.
## Static Plot
# ...characteristic vs. (presumed) rating...
# ...color&size mapped to own rating, grouped by ID
plot1 <- ggplot(df_what_look_for,
aes(x = value,
y = variable,
color = own_rating, # bubbels are always colord according to own whishes
size = own_rating,
group = iid)) + # identifier of observations across states
geom_jitter(alpha = 0.5, # to reduce overplotting: jitttering & alpha
width = 5) +
scale_color_viridis(option = "plasma", # use virdis' plasma scale
begin = 0.2, # limit range of used hues
name = "Own Rating") +
scale_size(guide = FALSE) + # no legend for size
labs(y = "", # no axis label
x = "Allocation of 100 Points", # x-axis label
title = "Importance of Characteristics for Potential Partner") +
theme_minimal() + # apply minimal theme
theme(panel.grid = element_blank(), # remove all lines of plot raster
text = element_text(size = 16)) # increase font size
## Animated Plot
plot1 +
transition_states(states = rating) # animate contrast subsets acc. to variable rating 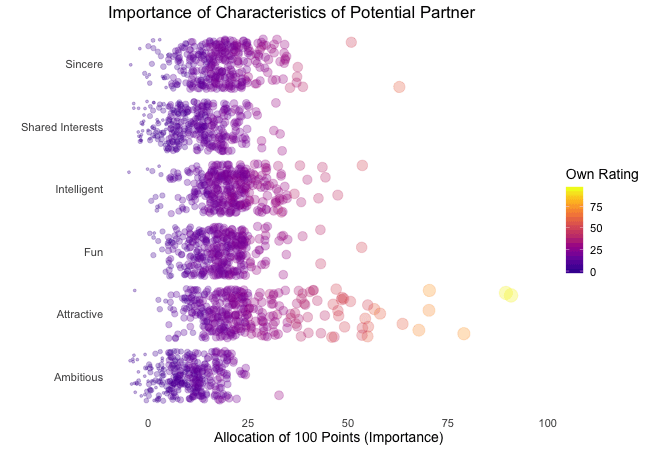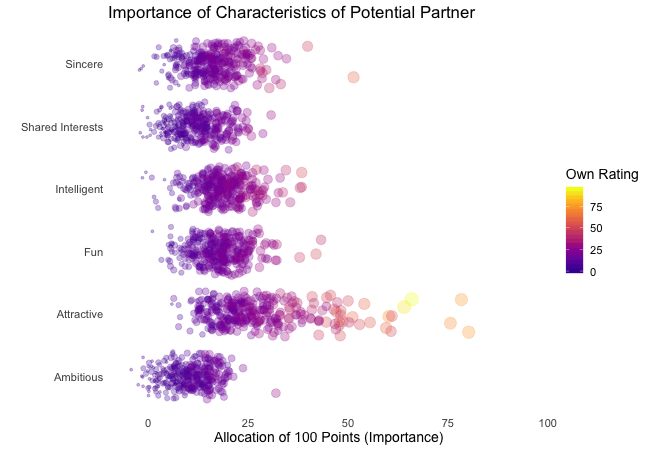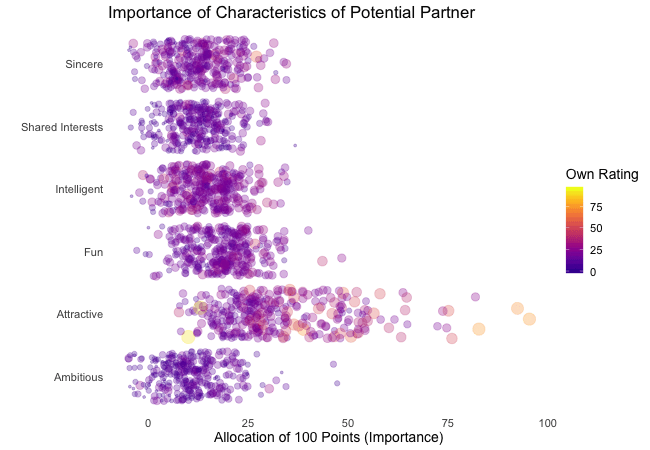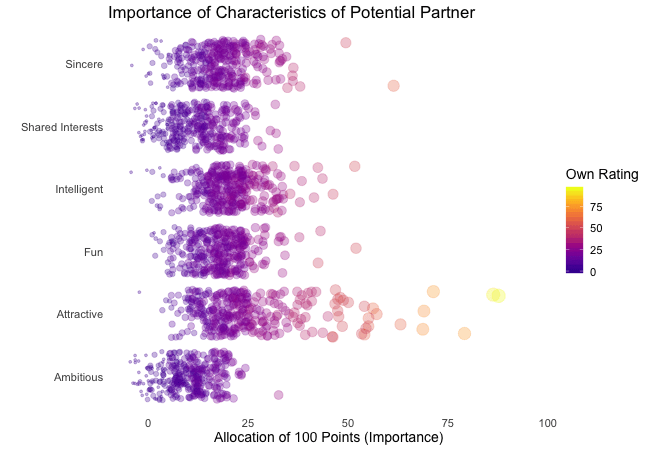
First off, if you’re a little confused which state is which, please be patient, we’ll explore dynamic labels in the section about ‚frame variables‘.
It’s apparent that different people look for different things in a partner. Yet attractiveness is often prioritized over other qualities. But the importance of attractiveness varies most strongly of all attributes between individuals. Interestingly, people are quite aware that their peer’s ratings might differ from their own views. Further, especially the collective presumptions (= the mean values) about others are not completely off, but of higher variance than the actual ratings.
So there is hope for all of us that somewhere out there somebody is looking for someone just as ambitious or just as intelligent as ourselves. However, it’s not always the inner values that count.
gganimate allows us to tailor the details of the animation according to our wishes. With the argument transition_length we can define the relative length of the transition from one to the other real subsets of data takes and with state_length how long, relatively speaking, each subset of original data is displayed. Only if the wrap argument is set to TRUE, the last frame will get morphed back into the first frame of the animation, creating an endless and seamless loop. Of course, the arguments of different transition functions may vary.
## Animated Plot
# ...replace default arguments
plot1 +
transition_states(states = rating,
transition_length = 3, # 3/4 of total time for transitions
state_length = 1, # 1/4 of time to display actual data
wrap = FALSE) # no endless loop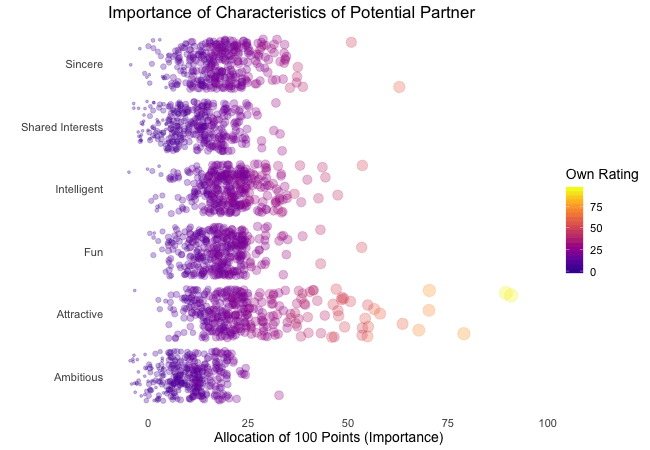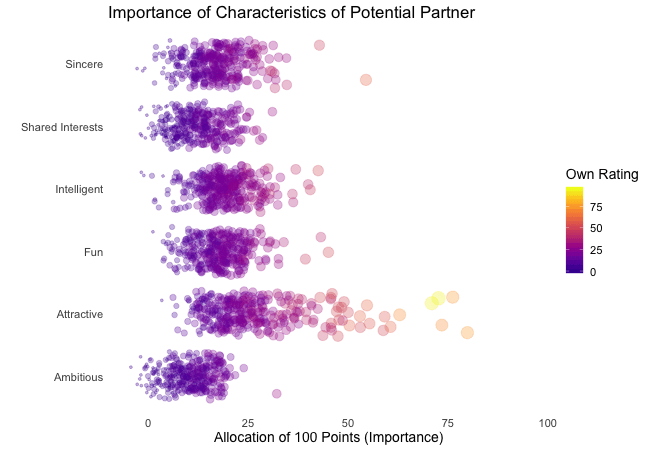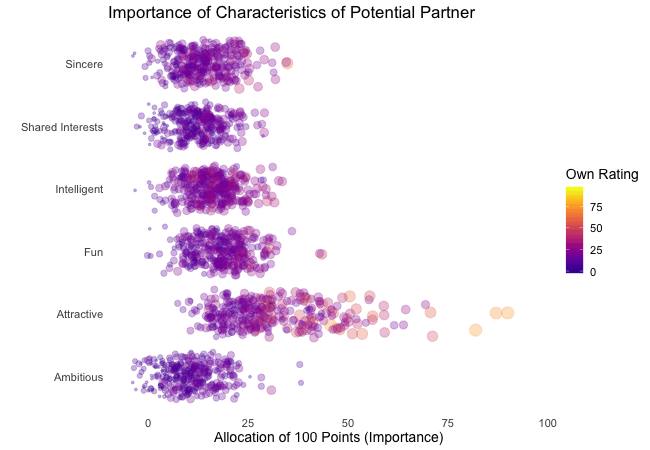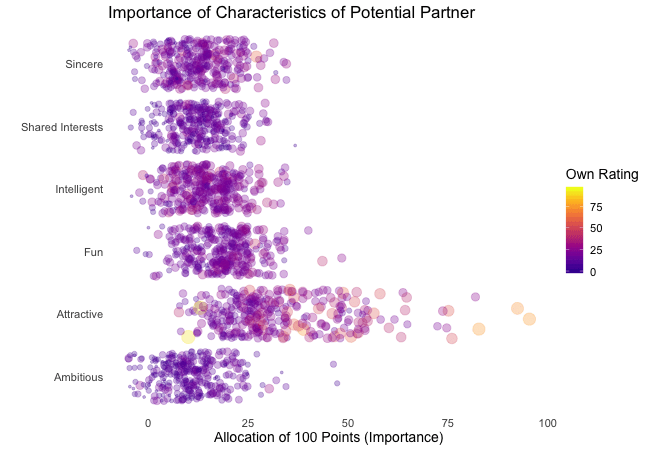
Styling transitions: ease_aes
As mentioned before, gganimate takes care of tweening and calculates additional data points to create smooth transitions between successively displayed points of actual input data. With ease_aes we can control which so-called easing function is used to ‚morph‘ original data points into each other. The default argument is used to declare the easing function for all aesthetics in a plot. Alternatively, easing functions can be assigned to individual aesthetics by name. Amongst others quadric, cubic , sine and exponential easing functions are available, with the linear easing function being the default. These functions can be customized further by adding a modifier-suffix: with -in the function is applied as-is, with -out the function is reversely applied with -in-out the function is applied as-is in the first half of the transition and reversed in the second half.
Here I played around with an easing function that models the bouncing of a ball.
## Animated Plot
# ...add special easing function
plot1 +
transition_states(states = rating) +
ease_aes("bounce-in") # bouncy easing function, as-is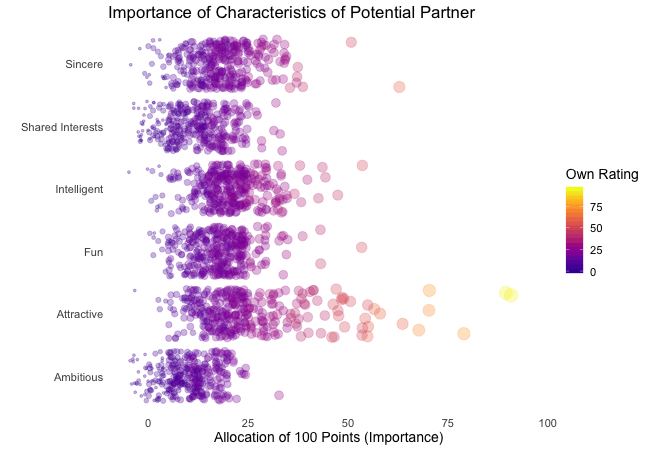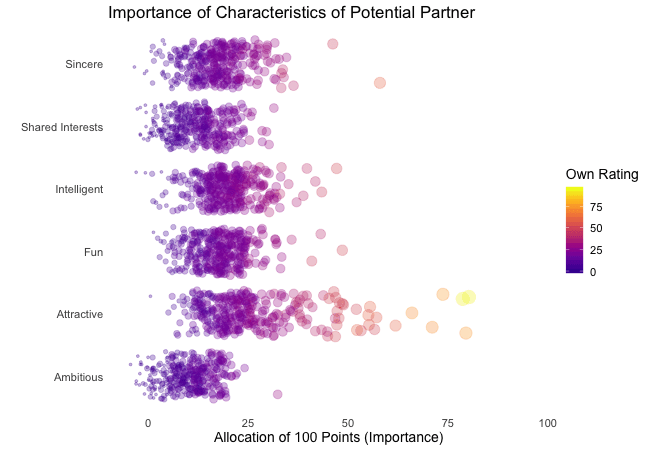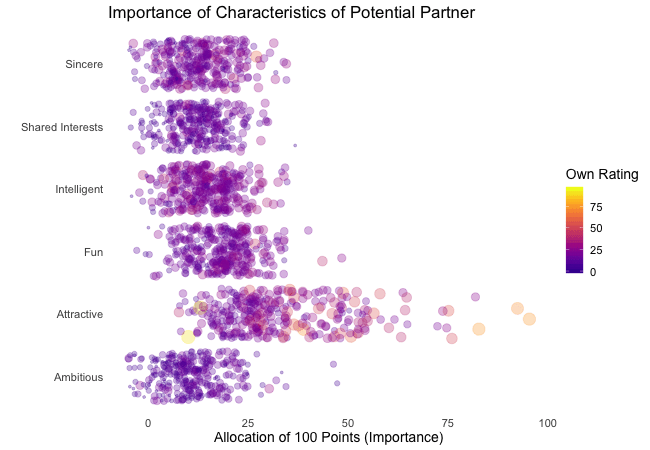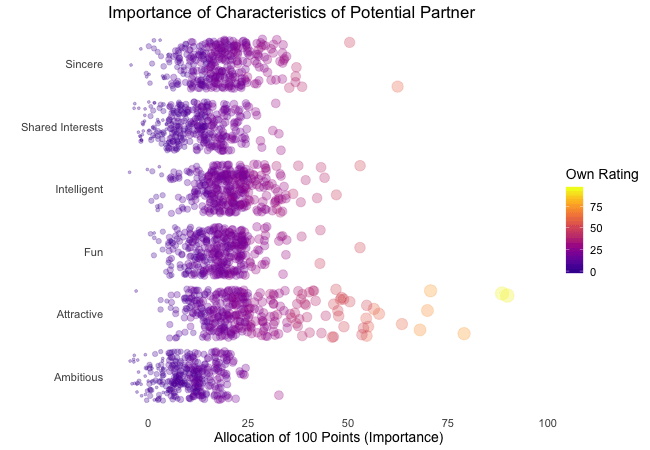
Dynamic labelling: {frame variables}
To ensure that we, mesmerized by our animations, do not lose the overview gganimate provides so-called frame variables that provide metadata about the animation as a whole or the previous/current/next frame. The frame variables – when wrapped in curly brackets – are available for string literal interpretation within all plot labels. For example, we can label each frame with the value of the states variable that defines the currently (or soon to be) displayed subset of actual data:
## Animated Plot
# ...add dynamic label: subtitle with current/next value of states variable
plot1 +
labs(subtitle = "{closest_state}") + # add frame variable as subtitle
transition_states(states = rating) 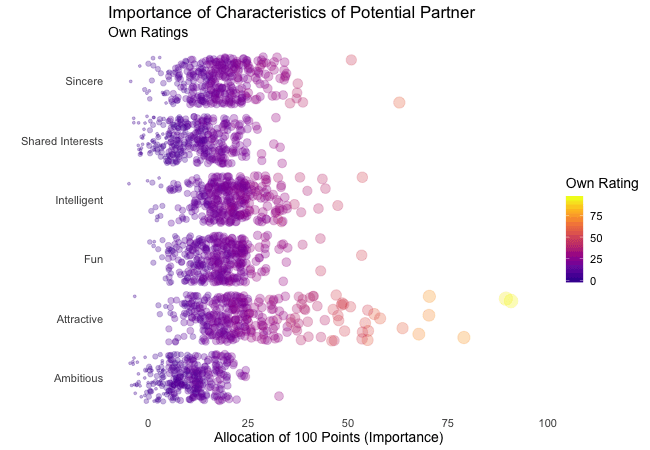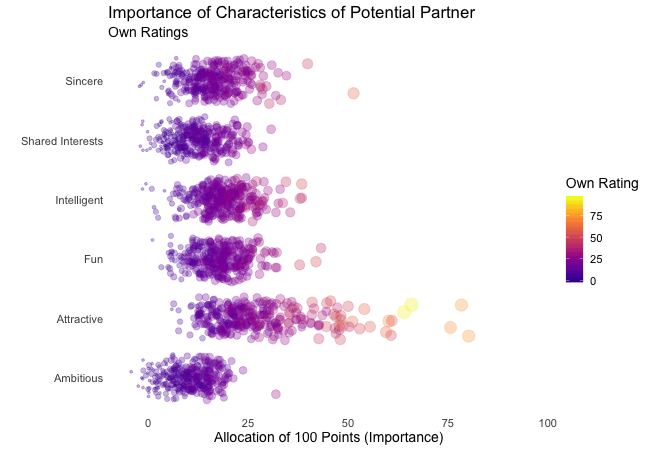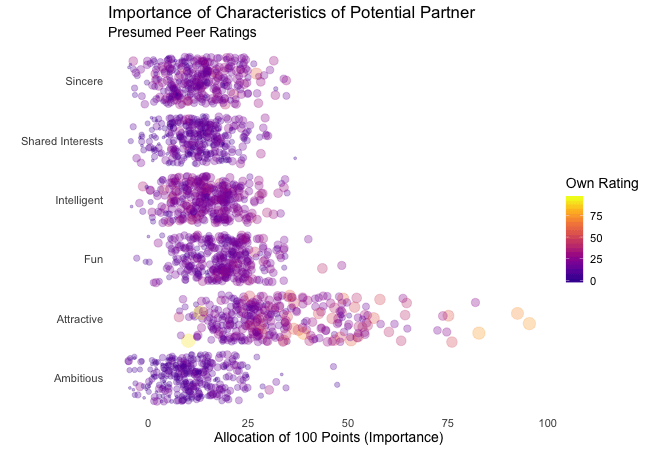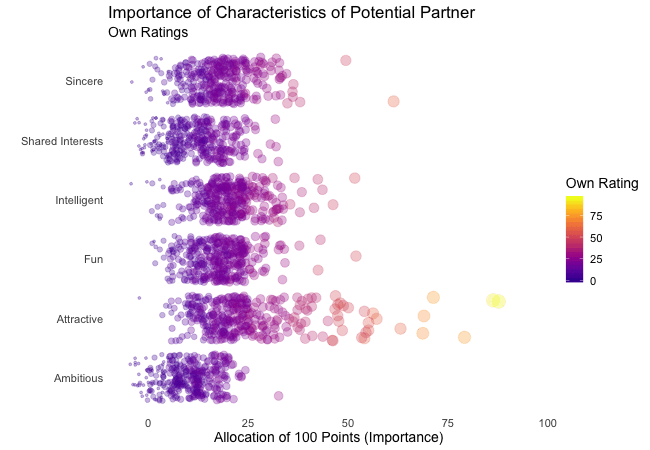
The set of available variables depends on the transition function. To get a list of frame variables available for any animation (per default the last one) the frame_vars() function can be called, to get both the names and values of the available variables.
Indicating previous data: shadow_*
To accentuate the interconnection of different frames, we can apply one of gganimates ’shadows‘. Per default shadow_null() i.e. no shadow is added to animations. In general, shadows display data points of past frames in different ways: shadow_trail() creates a trail of evenly spaced data points, while shadow_mark() displays all raw data points.
We’ll use shadow_wake() to create a little ‚wake‘ of past data points which are gradually shrinking and fading away. The argument wake_length allows us to set the length of the wake, relative to the total number of frames. Since the wakes overlap, the transparency of geoms might need adjustment. Obviously, for plots with lots of data points shadows can impede the intelligibility.
plot1B + # same as plot1, but with alpha = 0.1 in geom_jitter
labs(subtitle = "{closest_state}") +
transition_states(states = rating) +
shadow_wake(wake_length = 0.5) # adding shadow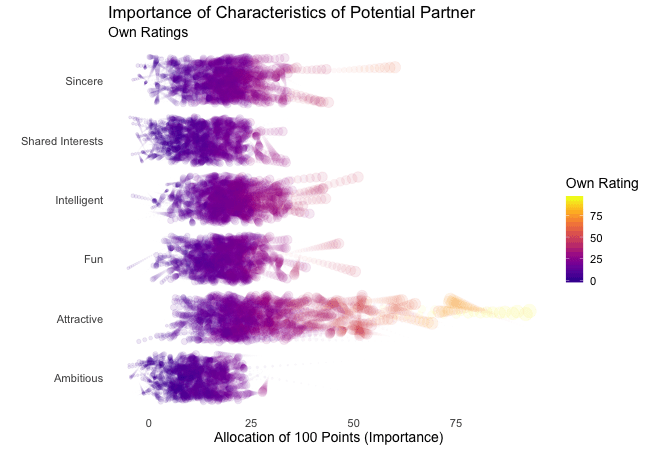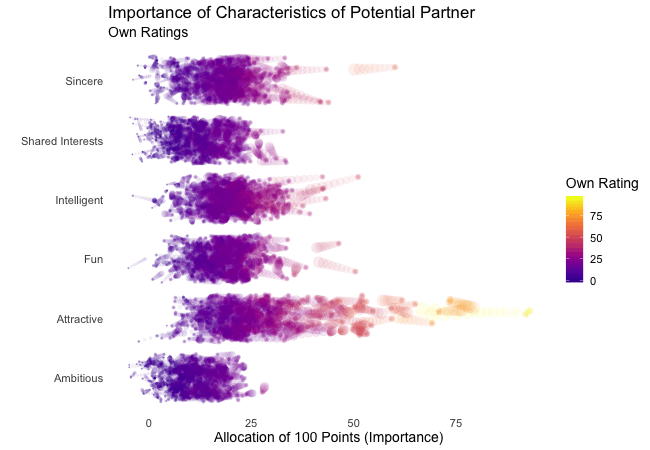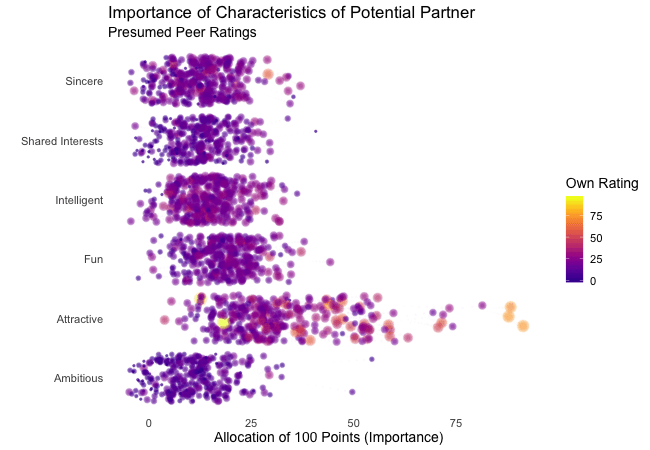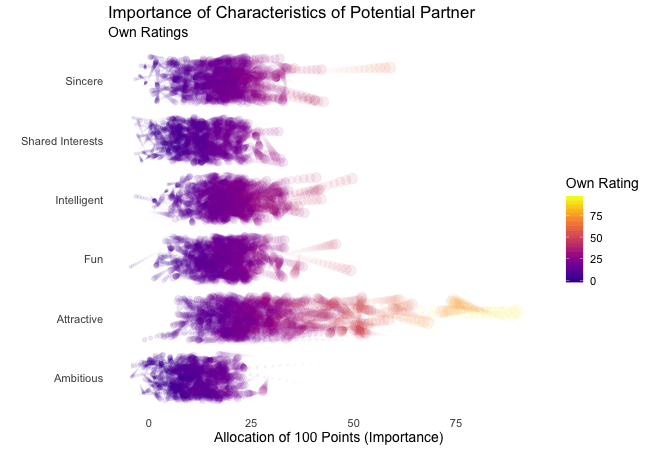
The benefits of transition_*
While I simply love the visuals of animated plots, I think they’re also offering actual improvement. I feel transition_states compared to facetting has the advantage of making it easier to track individual observations through transitions. Further, no matter how many subplots we want to explore, we do not need lots of space and clutter our document with thousands of plots nor do we have to put up with tiny plots.
Similarly, e.g. transition_reveal holds additional value for time series by not only mapping a time variable on one of the axes but also to actual time: the transition length between the individual frames displays of actual input data corresponds to the actual relative time differences of the mapped events. To illustrate this, let’s take a quick look at the ’success‘ of all the speed dates across the different speed dating events:
## Static Plot
# ... date of event vs. interest in second date for women, men or couples
plot2 <- ggplot(data = df_match,
aes(x = date, # date of speed dating event
y = count, # interest in 2nd date
color = info, # which group: women/men/reciprocal
group = info)) +
geom_point(aes(group = seq_along(date)), # needed, otherwise transition dosen't work
size = 4, # size of points
alpha = 0.7) + # slightly transparent
geom_line(aes(lty = info), # line type according to group
alpha = 0.6) + # slightly transparent
labs(y = "Interest After Speed Date",
x = "Date of Event",
title = "Overall Interest in Second Date") +
scale_linetype_manual(values = c("Men" = "solid", # assign line types to groups
"Women" = "solid",
"Reciprocal" = "dashed"),
guide = FALSE) + # no legend for linetypes
scale_y_continuous(labels = scales::percent_format(accuracy = 1)) + # y-axis in %
scale_color_manual(values = c("Men" = "#2A00B6", # assign colors to groups
"Women" = "#9B0E84",
"Reciprocal" = "#E94657"),
name = "") +
theme_minimal() + # apply minimal theme
theme(panel.grid = element_blank(), # remove all lines of plot raster
text = element_text(size = 16)) # increase font size
## Animated Plot
plot2 +
transition_reveal(along = date) 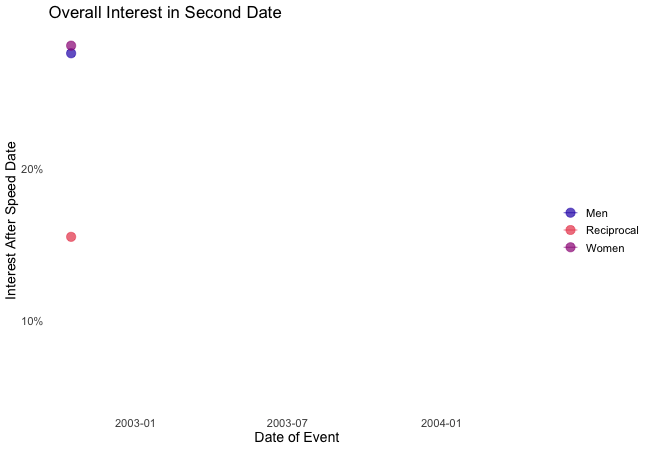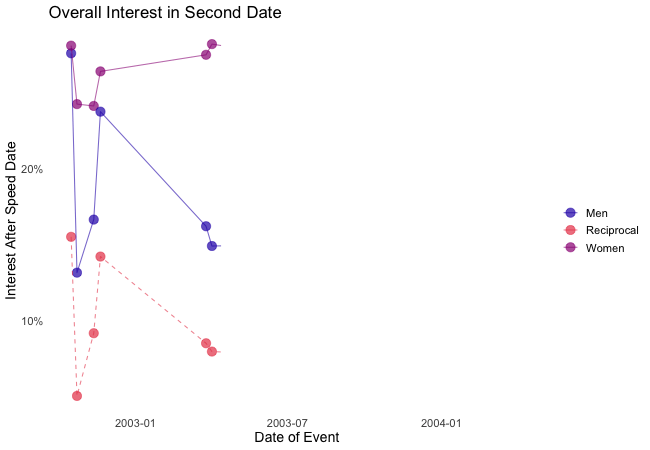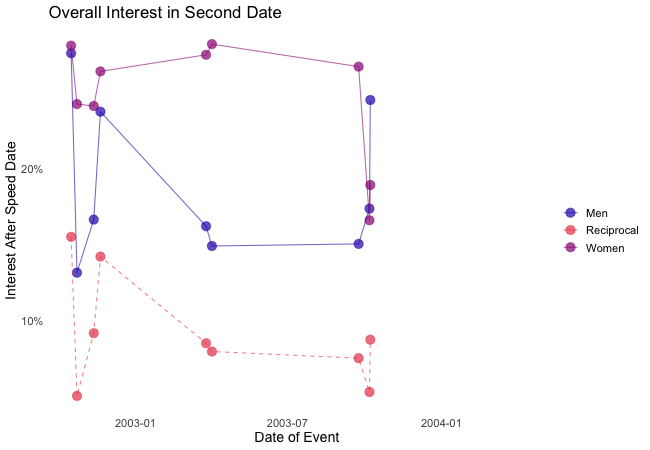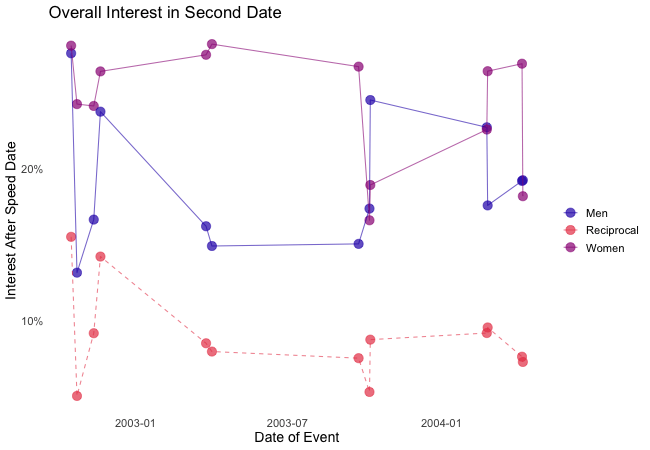
Displayed are the percentages of women and men who were interested in a second date after each of their speed dates as well as the percentage of couples in which both partners wanted to see each other again.
Most of the time, women were more interested in second dates than men. Further, the attraction between dating partners often didn’t go both ways: the instances in which both partners of a couple wanted a second date always were far more infrequent than the general interest of either men and women. While it’s hard to identify the most romantic time of the year, according to the data there seemed to be a slack in romance in early autumn. Maybe everybody still was heartbroken over their summer fling? Fortunately, Valentine’s Day is in February.
Another very handy option is transition_filter(), it’s a great way to present selected key insights of your data exploration. Here the animation browses through data subsets defined by a series of filter conditions. It’s up to you which data subsets you want to stage. The data is filtered according to logical statements defined in transition_filter(). All rows for which a statement holds true are included in the respective subset. We can assign names to the logical expressions, which can be accessed as frame variables. If the keep argument is set to TRUE, the data of previous frames is permanently displayed in later frames.
I want to explore, whether one’s own characteristics relate to the attributes one looks for in a partner. Do opposites attract? Or do birds of a feather (want to) flock together?
Displayed below are the importances the speed dating participants assigned to different attributes of a potential partner. Contrasted are subsets of participants, who were rated especially funny, attractive, sincere, intelligent or ambitious by their speed dating partners. The rating scale went from 1 = low to 10 = high, thus I assume value of >7 to be rather outstanding.
## Static Plot (without geom)
# ...importance ratings for different attributes
plot3 <- ggplot(data = df_ratings,
aes(x = variable, # different attributes
y = own_rating, # importance regarding potential partner
size = own_rating,
color = variable, # different attributes
fill = variable)) +
geom_jitter(alpha = 0.3) +
labs(x = "Attributes of Potential Partner", # x-axis label
y = "Allocation of 100 Points (Importance)", # y-axis label
title = "Importance of Characteristics of Potential Partner", # title
subtitle = "Subset of {closest_filter} Participants") + # dynamic subtitle
scale_color_viridis_d(option = "plasma", # use viridis scale for color
begin = 0.05, # limit range of used hues
end = 0.97,
guide = FALSE) + # don't show legend
scale_fill_viridis_d(option = "plasma", # use viridis scale for filling
begin = 0.05, # limit range of used hues
end = 0.97,
guide = FALSE) + # don't show legend
scale_size_continuous(guide = FALSE) + # don't show legend
theme_minimal() + # apply minimal theme
theme(panel.grid = element_blank(), # remove all lines of plot raster
text = element_text(size = 16)) # increase font size
## Animated Plot
# ...show ratings for different subsets of participants
plot3 +
geom_jitter(alpha = 0.3) +
transition_filter("More Attractive" = Attractive > 7, # adding named filter expressions
"Less Attractive" = Attractive <= 7,
"More Intelligent" = Intelligent > 7,
"Less Intelligent" = Intelligent <= 7,
"More Fun" = Fun > 7,
"Less Fun" = Fun <= 5) 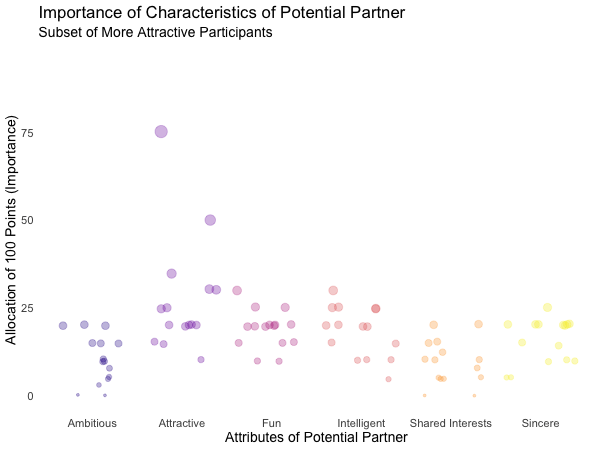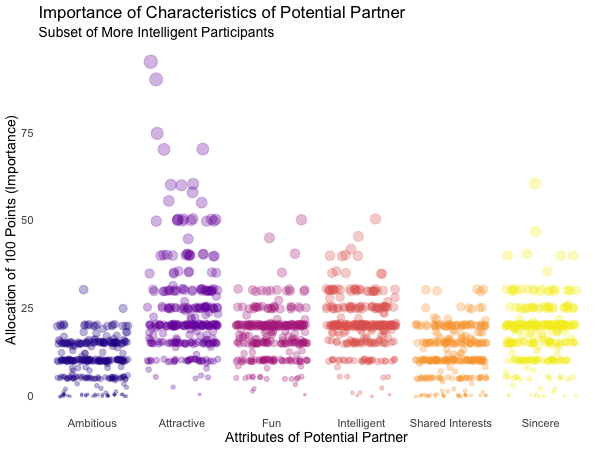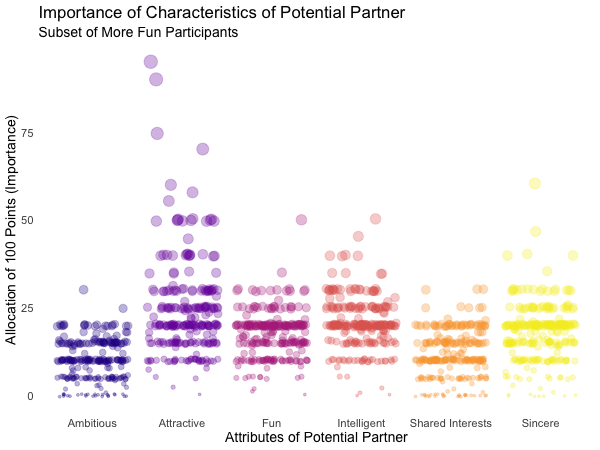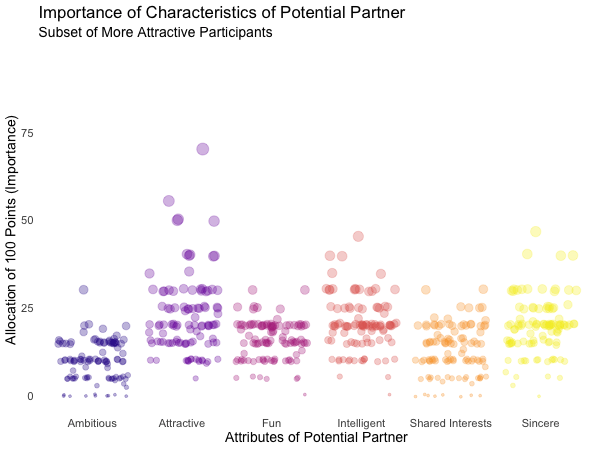
Of course, the number of extraordinarily attractive, intelligent or funny participants is relatively low. Surprisingly, there seem to be little differences between what the average low vs. high scoring participants look for in a partner. Rather the lower scoring group includes more people with outlying expectations regarding certain characteristics. Individual tastes seem to vary more or less independently from individual characteristics.
Styling the (dis)appearance of data: enter_* / exit_*
Especially if displayed subsets of data do not or only partially overlap, it can be favorable to underscore this visually. A good way to do this are the enter_*() and exit_*() functions, which enable us to style the entry and exit of data points, which do not persist between frames.
There are many combinable options: data points can simply (dis)appear (the default), fade (enter_fade()/exit_fade()), grow or shrink (enter_grow()/exit_shrink()), gradually change their color (enter_recolor()/exit_recolor()), fly (enter_fly()/exit_fly()) or drift (enter_drift()/exit_drift()) in and out.
We can use these stylistic devices to emphasize changes in the databases of different frames. I used exit_fade() to let further not included data points gradually fade away while flying them out of the plot area on a vertical route (y_loc = 100), data points re-entering the sample fly in vertically from the bottom of the plot (y_loc = 0):
## Animated Plot
# ...show ratings for different subsets of participants
plot3 +
geom_jitter(alpha = 0.3) +
transition_filter("More Attractive" = Attractive > 7, # adding named filter expressions
"Less Attractive" = Attractive <= 7,
"More Intelligent" = Intelligent > 7,
"Less Intelligent" = Intelligent <= 7,
"More Fun" = Fun > 7,
"Less Fun" = Fun <= 5) +
enter_fly(y_loc = 0) + # entering data: fly in vertically from bottom
exit_fly(y_loc = 100) + # exiting data: fly out vertically to top...
exit_fade() # ...while color is fading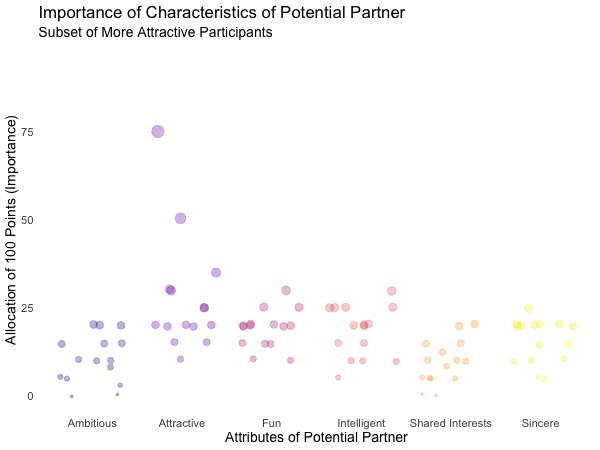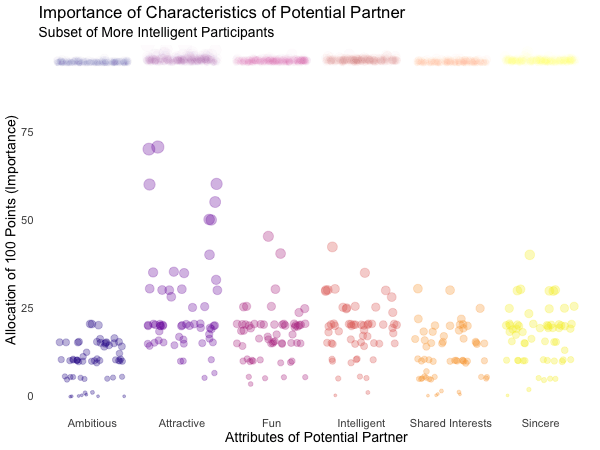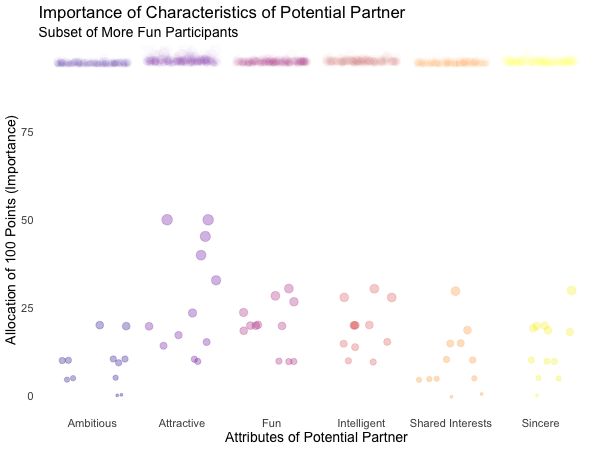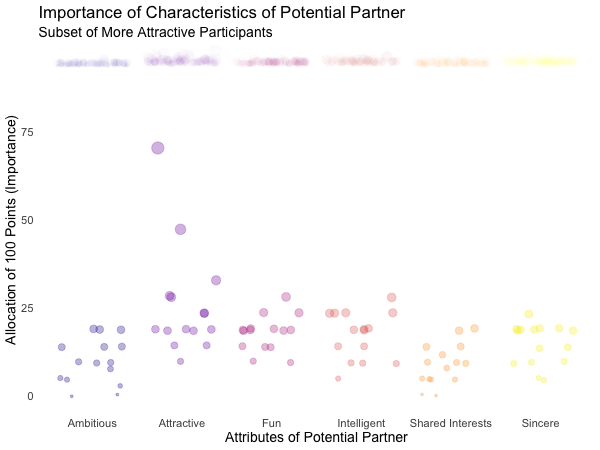
Finetuning and saving: animate() & anim_save()
Gladly, gganimate makes it very easy to finalize and save our animations. We can pass our finished gganimate object to animate() to, amongst other things, define the number of frames to be rendered (nframes) and/or the rate of frames per second (fps) and/or the number of seconds the animation should last (duration). We also have the option to define the device in which the individual frames are rendered (the default is device = “png”, but all popular devices are available). Further, we can define arguments that are passed on to the device, like e.g. width or height. Note, that simply printing an gganimateobject is equivalent to passing it to animate() with default arguments. If we plan to save our animation the argument renderer, is of importance: the function anim_save() lets us effortlessly save any gganimate object, but only so if it was rendered using one of the functions magick_renderer() or the default gifski_renderer().
The function anim_save()works quite straightforward. We can define filename and path (defaults to the current working directory) as well as the animation object (defaults to the most recently created animation).
# create a gganimate object
gg_animation <- plot3 +
transition_filter("More Attractive" = Attractive > 7,
"Less Attractive" = Attractive <= 7)
# adjust the animation settings
animate(gg_animation,
width = 900, # 900px wide
height = 600, # 600px high
nframes = 200, # 200 frames
fps = 10) # 10 frames per second
# save the last created animation to the current directory
anim_save("my_animated_plot.gif")Conclusion (and a Happy Valentine’s Day)
I hope this blog post gave you an idea, how to use gganimate to upgrade your own ggplots to beautiful and informative animations. I only scratched the surface of gganimates functionalities, so please do not mistake this post as an exhaustive description of the presented functions or the package. There is much out there for you to explore, so don’t wait any longer and get started with gganimate!
But even more important: don’t wait on love. The speed dating data shows that most likely there’s someone out there looking for someone just like you. So from everyone here at STATWORX: Happy Valentine’s Day!
## 8 bit heart animation
animation2 <- plot(data = df_eight_bit_heart %>% # includes color and x/y position of pixels
dplyr::mutate(id = row_number()), # create row number as ID
aes(x = x,
y = y,
color = color,
group = id)) +
geom_point(size = 18, # depends on height & width of animation
shape = 15) + # square
scale_color_manual(values = c("black" = "black", # map values of color to actual colors
"red" = "firebrick2",
"dark red" = "firebrick",
"white" = "white"),
guide = FALSE) + # do not include legend
theme_void() + # remove everything but geom from plot
transition_states(-y, # reveal from high to low y values
state_length = 0) +
shadow_mark() + # keep all past data points
enter_grow() + # new data grows
enter_fade() # new data starts without color
animate(animation2,
width = 250, # depends on size defined in geom_point
height = 250, # depends on size defined in geom_point
end_pause = 15) # pause at end of animation
Mein Blogbeitrag zielt auf Data Science Einsteiger ab, die vor der Wahl stehen, welche Programmiersprache sie als Erstes lernen wollen. Wir bei statworx arbeiten mit den zwei beliebtesten Sprachen R und Python. Beide Sprachen haben ihre Stärken und Schwächen, weshalb man idealerweise beide beherrschen sollte. Für den Einstieg empfehlen wir eine Sprache zu erlernen und sich dann in der anderen fortzubilden. Um die Entscheidung zu erleichtern, mit welcher Programmiersprache man beginnen möchte, stelle ich Euch beide vor und vergleiche sie anschließend miteinander.
Überblick R und Python
Sowohl Python als auch R sind Open-Source-Programmiersprachen. Das bedeutet, dass die Quellcodes öffentlich zugänglich sind und gratis verwendet werden können. Während Python eine General Purpose Programmiersprache (Allzwecksprache) ist, wurde R für statistische Analysen entwickelt. Daher weisen die Nutzer der Sprachen oftmals unterschiedliche Hintergründe auf. Verallgemeinernd kann man sagen, dass Softwareentwickler Python nutzen und Statistiker R.
| R | Python | |
|---|---|---|
| Veröffentlichung | 1993 | 1991 |
| Entwickler | R Core Team | Python Software Foundation |
| Package Management | CRAN | Conda (empfohlen für Einsteiger) |
Eine Fülle an Erweiterungen
Beide Sprachen verfügen über einen Grundstock an Funktionen, die mit Paketen (packages) erweitert werden können.
Das Comprehensive R Archive Network (CRAN) ist eine Plattform für R Pakete. Um ein Paket auf CRAN bereit zu stellen, müssen eine ganze Reihe an Richtlinien eingehalten werden. CRAN gewährleistet dadurch, dass alle Pakete, die dort zum Download zur Verfügung stehen auch tatsächlich funktionieren. Insgesamt stehen auf CRAN 10.000 Pakete zur Verfügung. Da R die Standard-Sprache für Statistiker ist, findet man in CRAN für fast jedes Problem im Bereich Statistik eine passende Lösung. Es ist also genau die richtige Anlaufstelle für die neuesten statistischen Methoden und Analysen.
Bei Python gibt es zwei Paket-Verwaltungsplattformen: conda und PyPI (Python Package Index). Auch für Python gibt es über 10.000 Pakete, die im Gegensatz zu R einen sehr breiten Anwendungsbereich abdecken. Da es zu Komplikationen kommen kann, wenn Python Pakete global installiert werden, nutzt man dafür virtuelle Umgebungen. Die sorgen für reibungslose Abläufe innerhalb der verschiedenen Pakete und bei Abhängigkeiten von Paket zu Paket. Für Anfänger ist es daher nicht so einfach, sich da zurecht zu finden.
Mit Hilfe von Paketen besteht die Möglichkeit in R Python Code auszuführen sowie vice versa. Falls dich das interessiert, check den Blogbeitrag von meinem Kollegen Manuel ab. Er stellt das Paket reticulate vor.
IDEs als Hilfestellung
Programmierer nutzen oftmals eine integrierte Entwicklungsumgebung (IDE), die ihnen die Arbeit durch kleine aber feine Hilfsmittel erleichtert.
Für R Nutzer hat sich RStudio als Standard-IDE durchgesetzt. Die IDE wird vom gleichnamigen Unternehmen vertrieben, das kommerziell hinter R steht. RStudio bietet nicht nur ein angenehmes Arbeitsumfeld, sondern entwickelt auch aktiv Pakete und Erweiterungen für die R Sprache. Vom RStudio-Team stammen beispielsweise wichtige Pakete wie tidyverse, packrat und devtools sowie beliebte Erweiterungen wie shiny (Dashboards) und RMarkdown (Berichte).
Python Nutzer haben die Wahl zwischen verschiedenen IDEs (PyCharm, Visual Studio Code, Spyder, …). Allerdings gibt es kein Unternehmen, das hinter Python steht und vergleichbar mit RStudio wäre. Dennoch werden dank der Bemühungen der riesigen Community und der Python Software Foundation ständig neue Erweiterungen für Python zusammengestellt.
Die Kunst der Datenvisualisierung
Die meist verwendeten Pakete für Datenvisualisierung mit Python sind matplotlib und seaborn. Dashboards lassen sich in Python mit dash erstellen.
Aber R hat bei der Datenvisualisierung einen Trumpf im Ärmel: Das Paket ggplot2, das auf dem Buch The Grammar of Graphics von Leland Wilkinson basiert. Mit diesem Paket kannst Du ansprechende und maßgeschneiderte Grafiken erstellen, die Du wiederum auf Dashboards mit Hilfe von shiny für Andere zugänglich machen kannst.
Beide Programmiersprachen bieten die Möglichkeit, schöne Grafiken leicht zu erstellen. Trotzdem überzeugt das R Paket ggplot2 mit seiner Flexibilität und seinen visuellen Möglichkeiten.
Pluspunkte für Lesbarkeit
Python wurde nach dem Motto Readability counts konzipiert. Somit können auch Leute, die nicht mit der Programmiersprache vertraut sind, interpretieren was im Code gemacht wird.
Das ist in R Code eher nicht der Fall. Die Sprache ist weniger intuitiv aufgebaut als Python. Aufgrund der guten Lesbarkeit bietet Python daher einen leichteren Einstieg ins Programmieren.
Schnelligkeit in verschiedenen Observationsgrößen
Als nächstes vergleiche ich, wie lange es dauert in R und Python einen simulierten Datensatz zu erstellen. Für eine faire Gegenüberstellung sollten die Bedingungen möglichst gleich sein. Die Daten werden mit den Paketen Xy und XyPy in R und Python respektive simuliert. Für die Zeitmessung habe ich microbenchmark in R und timeit in Python benutzt. Um die Simulation schnellstmöglich zu generieren, wird der Prozess parallelisiert auf acht Kernen (R: parallel, Python: multiprocessing).
Für das Experiment wird ein Datensatz mit 100 Observationen und 50 Variablen 100 Mal simuliert. Die Zeit, die der Rechner benötigt, um die Simulation durchzuführen, wird für jede Simulation einzeln gemessen. Und das wird dann für 1.000, 10.000, 100.000 und 1.000.000 Observationen wiederholt.
Die R und Python Code Snippets sind unten abgebildet.
# R
# devtools::install_github("andrebleier/Xy")
# install.packages("parallel")
# install.packages("microbenchmark")
# Load packages
library(Xy)
library(microbenchmark)
library(parallel)
# Extract function definition from for loop
sim_this <- function(n_sim) {
sim <- microbenchmark(Xy(n = n_sim,
numvars = c(50,0),
catvars = 0),
times = 100, unit = "s")
data.frame(n = n_sim,
mean = summary(sim)[, 4])
}
# Time measurement for different number of simulations
n_sim <- c(1e2, 1e3, 1e4, 1e5, 1e6)
sim_in_r <- data.frame(n = rep(0, length(n_sim)),
t = rep(0, length(n_sim)))
for(i in 1:length(n_sim)){
out <- mclapply(n_sim[i],
FUN = sim_this,
mc.cores = 8)
sim_in_r[i, 1] <- out[[1]][1]
sim_in_r[i, 2] <- out[[1]][2]
}
# Python
# In terminal: pip install xypy
import multiprocessing as mp
import numpy as np
import timeit
from xypy import Xy
# Predefine function of interest
def sim_this(n_sim):
return(timeit.timeit( lambda: Xy(n = int(n_sim),
numvars = [50, 0],
catvars = [0, 0],
weights = [5, 10],
stn = 4.0,
cor = [0, 0.1],
interactions = 1,
noisevars = 5), number = 100))
# Paralleled computation
pool = mp.Pool(processes = 8)
n_sim = np.array([1e2, 1e3, 1e4, 1e5, 1e6])
results = [pool.map(sim_this, n_sim)]
Die durchschnittliche Dauer, sortiert nach Datensatzgröße, wird für R und Python im unteren Plot dargestellt. Die X-Achse wird hier auf einer logarithmischen Skala mit Basis 10 dargestellt, um die Grafik übersichtlicher zu machen.
Während R bei einer Datensatzgröße von 100 und 1.000 Observationen etwas schneller ist, hängt Python R bald darauf deutlich ab.
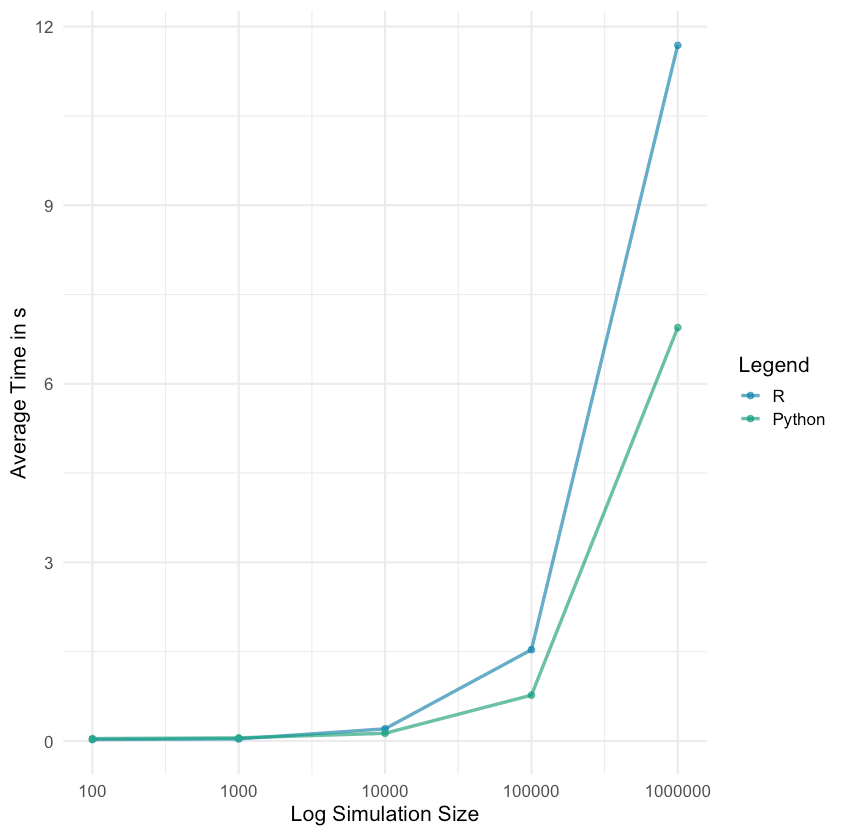
Für weitere Vergleiche kann ich die folgenden STATWORX Blogbeiträge empfehlen: pandas vs. data.table und pandas vs. data.table part 2, dabei wird der Fokus auf Datenmanipulation gelegt.
Der Standard bei Deep Learning
Wenn Dich vor allem Deep Learning Methoden interessieren, eignet sich Python als Sprache besser. Die meisten Deep Learning Bibliotheken wurden in Python geschrieben und implementiert.
Auch in R ist Deep Learning möglich, aber die R Deep Learning Community ist deutlich kleiner. Implementationen wie Keras und TensorFlow lassen sich zwar auch in R aufrufen, dies läuft dann aber über Pakete von Drittanbietern. Die Pakete bieten daher nicht die volle Flexibilität für die Nutzer, z.B. sind nicht alle TensorFlow Funktionen erhältlich. Zu dem kommt der Aspekt der Schnelligkeit. Deep Learning mit Python ist schneller als mit R.
Umfrage in der Community: Wie ticken die Anwender?
Als angehende Data Scientists ist Kaggle eine wichtige Plattform für Euch. Dort kann man an spannenden Machine Learning Wettkämpfen teilnehmen, selbst experimentieren und aus den Erfahrungen der Community lernen.
2018 hat Kaggle eine Machine Learning & Data Science Umfrage durchgeführt. Die Umfrage war zwei Wochen lang online und es gingen insgesamt 23.859 Antworten ein. Aus den Ergebnissen dieser Umfrage habe ich verschiedene Plots erstellt, aus denen sich einige interessante Schlüsse im Hinblick auf mein Blogthema ziehen lassen. Der Code zu den einzelnen Plots ist öffentlich zugänglich auf Github.
Exkurs: Python & R im Vergleich zu anderen Sprachen
Bevor wir uns auf R und Python stürzen, schauen wir uns an, wie die beiden im Vergleich zu anderen Programmiersprachen abschneiden. Jeder Umfragenteilnehmer gab an, welche Sprache er vorrangig benutzt. Im unteren Plot wurde nach Sprache aggregiert und das Ergebnis lautet: Die große Mehrzahl der Teilnehmer benutzt vor allem Python! Gefolgt von R auf dem zweiten Platz. In dieser Umfrage unterscheiden wir nicht zwischen den Arbeitsbereichen, weshalb Python – als General Purpose Programmiersprache – vermutlich so stark hervorsticht.
Die Gegenüberstellung R & Python
Im direkten Vergleich zwischen R und Python sieht man, dass sehr viele R-Nutzer auch Python benutzen. Wohingegen die Python-Nutzer oftmals ausschließlich mit Python arbeiten.
Wenn man die Nutzung der Sprachen nach Arbeitsbereich vergleicht, sieht man eine klare Dominanz von Python. In allen Arbeitsfeldern, bis auf Statistiker, wird mehrheitlich Python benutzt.

Die Teilnehmer wurden außerdem gefragt: Welche Sprache empfiehlst Du angehenden Data Scientists zuerst zu lernen? Die Antworten auf die Frage sind in der unteren Tabelle zusammengefasst.
| Sprache | Empfehlung | Nutzer | Differenz |
|---|---|---|---|
| Python | 14.181 | 8.180 | 6.001 |
| R | 2.342 | 2.046 | 296 |
| SQL | 914 | 1.211 | -297 |
| C++ | 339 | 739 | -400 |
| Matlab | 256 | 355 | -99 |
| Java | 184 | 903 | -719 |
| Scala | 74 | 106 | -32 |
| Javascript | 72 | 408 | -336 |
| SAS | 69 | 228 | -159 |
| VBA | 38 | 135 | -97 |
| Go | 26 | 46 | -20 |
| Other | 161 | 117 | 44 |
Wenn man die Anzahl Empfehlungen und die Anzahl Nutzer vergleicht, dann sieht man, dass R und Python die einzigen Sprachen sind, die eine positive Differenz aufweisen.
Auch bei dieser Frage liegt Python (14.181) wieder weit vor R (2.342).
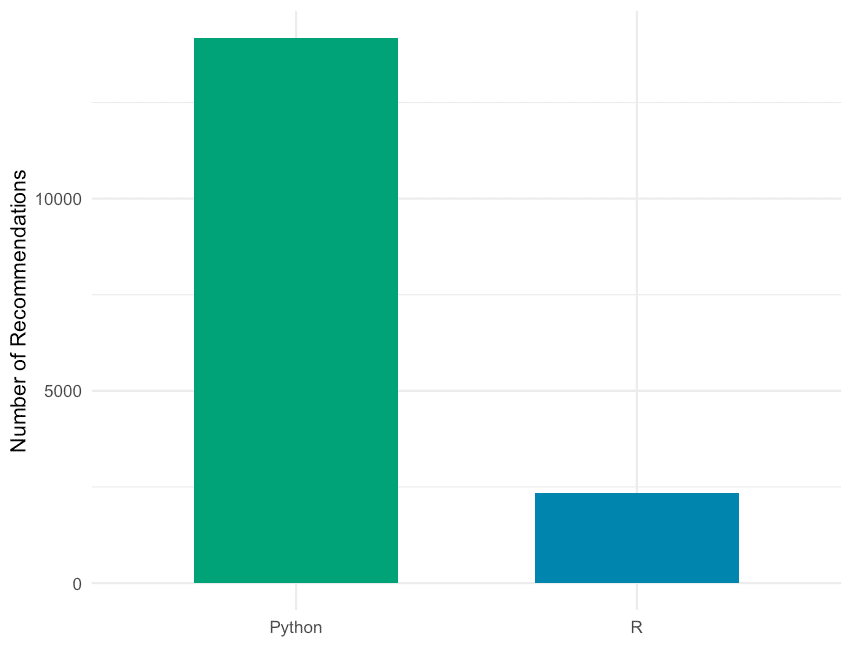
Fazit
Eine Sache vorweg: beide Sprachen sind sehr mächtig. Daher kann man keine falsche Wahl treffen! Die Wahl der Sprache hängt davon ab, welche Projekte man verwirklichen möchte.
Als universelle Programmiersprache ist Python für diverse Anwendungsgebiete geeignet. Weshalb ich Dir grundsätzlich empfehle mit Python anzufangen. Falls aber statistische Auswertungen oder Datenvisualisierungen bei Deinen Projekten im Vordergrund stehen, hat R gegenüber Python einen Vorteil.
Wie schon erwähnt haben beide Sprachen ihre Vor- und Nachteile. Als fortgeschrittener Data Scientist solltest Du idealerweise beide Sprachen beherrschen.
Ich hoffe, dass Dir dieser Beitrag bei der Suche nach dem richtigen Einstieg in die Data Science Welt weiterhilft.
Happy Coding!
Falls Du Interesse an Schulungen hast, kannst du dir gerne unter AI Academy unsere Kurse durchschauen.
Referenzen
- https://cran.r-project.org/
- https://www.python.org/
- https://wiki.c2.com/?PythonPhilosophy
- https://plot.ly/
- https://www.rstudio.com/
- https://pipenv.readthedocs.io/en/latest/
- https://conda.io/en/latest/
- https://www.kaggle.com
- https://www.springer.com/us/book/9780387245447
- https://www.statworx.com/ai-academy/
Intro
Informationen sind überall im Internet zu finden. Leider ist es schwierig, programmatisch auf einige davon zuzugreifen. Zwar bieten viele Websites eine API an, doch sind diese oft teuer oder haben sehr strenge Tarifbeschränkungen, selbst wenn Sie an einem Open-Source- und/oder nicht kommerziellen Projekt oder Produkt arbeiten.
Hier kann Web Scraping ins Spiel kommen. Wikipedia definiert Web Scraping wie folgt:
Web scraping, web harvesting, or web data extraction data scraping used for extracting data from websites. Web scraping software may access the World Wide Web directly using the Hypertext Transfer Protocol [HTTP], or through a web browser.
“Web scraping” wikipedia.org
In der Praxis umfasst Web Scraping jede Methode, die es Programmierer:innen ermöglicht, programmatisch und damit (halb-)automatisch auf den Inhalt einer Website zuzugreifen.
Hier sind drei Ansätze (d.h. Python-Bibliotheken) für Web Scraping, die zu den beliebtesten gehören:
- Senden einer HTTP-Anfrage, üblicherweise über Requests, an eine Webseite und anschließendes Parsen des zurückgegebenen HTML (üblicherweise mit BeautifulSoup), um auf die gewünschten Informationen zuzugreifen. Typischer Use Case: Standard-Web-Scraping-Problem, siehe Fallstudie in diesem Beitrag.
- Verwendung von Tools, die normalerweise für automatisierte Softwaretests verwendet werden, hauptsächlich Selenium, um programmatisch auf den Inhalt einer Website zuzugreifen. Typischer Anwendungsfall: Websites, die Javascript verwenden oder anderweitig nicht direkt über HTML zugänglich sind.
- Scrapy, das eher als allgemeines Web-Scraping-Framework betrachtet werden kann, mit dem Spiders erstellt und Daten von verschiedenen Websites gescraped werden können, wobei Wiederholungen minimiert werden. Typischer Anwendungsfall: Scraping von Amazon-Rezensionen.
Obwohl man Daten auch mit jeder anderen Programmiersprache scrapen könnte, wird Python aufgrund seiner einfachen Syntax und der großen Vielfalt an Bibliotheken, die für Scraping-Zwecke in Python zur Verfügung stehen, am häufigsten verwendet.
Nach dieser kurzen Einführung geht es in diesem Beitrag um einige ethische Aspekte des Web-Scraping, gefolgt von allgemeinen Informationen über die Bibliotheken, die in diesem Beitrag verwendet werden. Schließlich wird alles, was wir bisher gelernt haben, auf eine Fallstudie angewandt, in der wir die Daten aller Unternehmen im Portfolio von Sequoia Capital, einer der bekanntesten VC-Firmen in den USA, erfassen werden. Nach einer Überprüfung der Website und der robots.txt scheint das Scraping des Sequoia-Portfolios erlaubt zu sein; wie ich das herausgefunden habe, erfahren Sie im Abschnitt über robots.txt und in der Fallstudie.
Im Rahmen dieses Blogbeitrags können wir uns nur eine der drei oben genannten Methoden ansehen. Da die Standardkombination von Requests + BeautifulSoup im Allgemeinen am flexibelsten und am einfachsten zu handhaben ist, werden wir sie in diesem Beitrag ausprobieren. Beachten Sie, dass die oben genannten Tools sich nicht gegenseitig ausschließen; Sie könnten z.B. einen HTML-Text mit Scrapy oder Selenium erhalten und ihn dann mit BeautifulSoup parsen.
Web Scraping Ethik
Zwei Faktoren, die bei der Durchführung von Web-Scraping äußerst wichtig sind, sind Ethik und Rechtmäßigkeit. Ich bin kein Jurist, und die spezifischen Gesetze variieren je nach Land beträchtlich, aber im Allgemeinen fällt Web Scraping in eine Grauzone, d.h. es ist in der Regel nicht streng verboten, aber auch nicht generell legal (d.h. nicht unter allen Umständen legal). Das hängt in der Regel von den Daten ab, die Sie auslesen.
Im Allgemeinen können Websites Ihre IP-Adresse sperren, wenn Sie etwas auslesen, was nicht erwünscht ist. Wir hier bei STATWORX dulden keine illegalen Aktivitäten und empfehlen Ihnen, immer ausdrücklich nachzufragen, wenn Sie sich nicht sicher sind, ob das Scrapen von Daten zulässig ist. Hierfür ist der folgende Abschnitt sehr nützlich.
robots.txt verstehen
Der robot exclusion standard ist ein Protokoll, das von Web-Crawlern (z. B. von den großen Suchmaschinen, d. h. hauptsächlich Google) ausdrücklich gelesen wird und ihnen mitteilt, welche Teile einer Website vom Crawler indiziert werden dürfen und welche nicht. Im Allgemeinen sind Crawler oder Scraper nicht gezwungen, die in einer robots.txt festgelegten Einschränkungen zu befolgen, aber es wäre höchst unmoralisch (und potenziell illegal), dies nicht zu tun.
Das folgende Beispiel zeigt eine robots.txt-Datei von Hackernews, einem sozialen Newsfeed, der von YCombinator betrieben wird und bei vielen Menschen in Startups beliebt ist.
In der robots.txt von Hackernews ist festgelegt, dass alle User Agents (daher der Platzhalter *) auf alle URLs zugreifen dürfen, mit Ausnahme der URLs, die ausdrücklich verboten sind. Da nur bestimmte URLs verboten sind, ist damit implizit alles andere erlaubt. Eine Alternative wäre, alles auszuschließen und dann explizit nur bestimmte URLs anzugeben, auf die Crawler oder andere Bots zugreifen können.
Beachten Sie auch die Crawl-Verzögerung von 30 Sekunden, was bedeutet, dass jeder Bot nur eine Anfrage alle 30 Sekunden senden sollte. Es ist im Allgemeinen eine gute Praxis, Ihren Crawler oder Scraper in regelmäßigen (ziemlich großen) Abständen schlafen zu lassen, da zu viele Anfragen Websites zum Absturz bringen können, selbst wenn sie von menschlichen Nutzenden stammen.
Wenn man sich die robots.txt von Hackernews ansieht, ist es auch ziemlich logisch, warum sie einige bestimmte URLs nicht zulassen: Sie wollen nicht, dass Bots sich als User ausgeben, indem sie z.B. Themen einreichen, abstimmen oder antworten. Alles andere (z.B. das Scrapen von Themen und deren Inhalten) ist erlaubt, solange die Crawl-Verzögerung eingehalten wird. Das macht Sinn, wenn man die Aufgabe von Hackernews bedenkt, die hauptsächlich darin besteht, Informationen zu verbreiten. Übrigens bieten sie auch eine API an, die recht einfach zu benutzen ist. Wenn Sie also wirklich Informationen von HN benötigen, würden Sie einfach ihre API benutzen.
In der Gist unten finden Sie die robots.txt von Google, die (natürlich) viel restriktiver ist als die von Hackernews. Schauen Sie selbst nach, denn sie ist viel länger als unten gezeigt, aber im Wesentlichen ist es Bots nicht erlaubt, eine Suche bei Google durchzuführen, wie in den ersten beiden Zeilen angegeben. Nur bestimmte Teile einer Suche sind erlaubt, wie „about“ und „static“. Wenn eine allgemeine URL verboten ist, wird sie überschrieben, wenn eine spezifischere URL erlaubt wird (z.B. wird das Verbot von /search durch das spezifischere Zulassen von /search/about überschrieben).
Im Folgenden werden wir einen Blick auf die spezifischen Python-Pakete werfen, die im Rahmen dieser Fallstudie verwendet werden, nämlich Requests und BeautifulSoup.
Requests
Requests ist eine Python-Bibliothek, mit der man auf einfache Weise HTTP-Anfragen stellen kann. Im Allgemeinen hat Requests zwei Hauptanwendungsfälle: Anfragen an eine API und das Abrufen von Roh-HTML-Inhalten von Websites (d.h. Scraping).
Wenn Sie eine Anfrage senden, sollten Sie immer den Statuscode überprüfen (insbesondere beim Scraping), um sicherzustellen, dass Ihre Anfrage erfolgreich bearbeitet wurde. Eine nützliche Übersicht über die Statuscodes finden Sie hier. Im Idealfall sollte Ihr Statuscode 200 sein (was bedeutet, dass Ihre Anfrage erfolgreich war). Der Statuscode kann Ihnen auch Auskunft darüber geben, warum Ihre Anfrage nicht zugestellt werden konnte, z.B. weil Sie zu viele Anfragen gesendet haben (Statuscode 429) oder die berüchtigte Seite nicht gefunden wurde (Statuscode 404).
Use Case 1: API Requests
Die obige Gist zeigt eine einfache API-Anfrage, die an die NYT-API gerichtet ist. Wenn Sie diese Anfrage auf Ihrem eigenen Rechner replizieren möchten, müssen Sie zunächst ein Konto auf der Seite NYT Dev erstellen und dann den erhaltenen Schlüssel der Konstante KEY zuweisen.
Die Daten, die Sie von einer REST-API erhalten, liegen im JSON-Format vor, das in Python als eine dict-Datenstruktur dargestellt wird. Daher müssen Sie diese Daten noch ein wenig „parsen“, bevor Sie sie in einem Tabellenformat haben, das z.B. in einer CSV-Datei dargestellt werden kann, d.h. Sie müssen auswählen, welche Daten für Sie relevant sind.
Use Case 2: Scraping
Die folgenden Zeilen fragen den HTML-Code der Wikipedia-Seite zum Thema Web Scraping ab. Das Statuscode-Attribut des Response-Objekts enthält den mit der Anfrage verbundenen Statuscode.
Nach dem Ausführen dieser Zeilen haben Sie immer noch nur den rohen HTML-Code mit allen enthaltenen Tags. Dies ist in der Regel nicht sehr nützlich, da wir beim Scraping mit Requests meist nur nach bestimmten Informationen und Text suchen, da menschliche Lesende nicht an HTML-Tags oder anderen Markups interessiert sind. An dieser Stelle kommt BeautifulSoup ins Spiel.
BeautifulSoup
BeautifulSoup ist eine Python-Bibliothek, die zum Parsen von Dokumenten (d.h. hauptsächlich HTML- oder XML-Dateien) verwendet wird. Die Verwendung von Requests, um den HTML-Code einer Seite zu erhalten, und das anschließende Parsen der gesuchten Informationen mit BeautifulSoup aus dem rohen HTML-Code ist der Quasi-Standard für Web-Scraping, der von Python-Programmierern häufig für einfache Aufgaben verwendet wird.
Um auf die obige Gist zurückzukommen, würde das Parsen des von Wikipedia zurückgegebenen HTML-Rohmaterials für die Web-Scraping-Site ähnlich wie unten aussehen.
In diesem Fall extrahiert BeautifulSoup alle Überschriften, d.h. alle Überschriften im Abschnitt „Inhalt“ oben auf der Seite. Probieren Sie es selbst aus!
Wie Sie unten sehen, können Sie das Klassenattribut eines HTML-Elements leicht mit dem Inspektor eines beliebigen Webbrowsers finden.
Diese Art des Abgleichs ist (meiner Meinung nach) eine der einfachsten Möglichkeiten, BeautifulSoup zu verwenden: Sie geben einfach das HTML-Tag (in diesem Fall span) und ein anderes Attribut des Inhalts an, das Sie finden wollen (in diesem Fall ist dieses andere Attribut class). Auf diese Weise können Sie beliebige Abschnitte fast jeder Webseite abgleichen. Für kompliziertere Übereinstimmungen können Sie auch reguläre Ausdrücke (REGEX) verwenden.
Sobald Sie die Elemente haben, aus denen Sie den Text extrahieren möchten, können Sie auf den Text zugreifen, indem Sie deren Textattribut auslesen.
Inspector
Als kurzer Exkurs ist es wichtig, eine Einführung in die in Chrome zu geben (sie sind in jedem Browser verfügbar, ich habe mich nur für Chrome entschieden), mit denen Sie den Inspektor verwenden können, der Ihnen Zugriff auf den HTML-Code einer Website gibt und mit dem Sie auch Attribute wie den XPath- und CSS-Selektor kopieren können. All dies kann beim Scraping-Prozess hilfreich oder sogar notwendig sein (vor allem bei der Verwendung von Selenium). Der Arbeitsablauf in der Fallstudie sollte Ihnen einen grundlegenden Eindruck davon vermitteln, wie Sie mit dem Inspector arbeiten können. Ausführlichere Informationen über den Inspector finden Sie auf der oben verlinkten offiziellen Google-Website, die zahlreiche Informationen enthält.
Abbildung 2 zeigt die grundlegende Schnittstelle des Inspektors in Chrome.
Fallstudie: Sequoia Capital
Eigentlich wollte ich diese Fallstudie zunächst mit der New York Times durchführen, da sie über eine API verfügt und somit die von der API erhaltenen Ergebnisse mit den Ergebnissen des Scrapings hätten verglichen werden können. Leider haben die meisten Nachrichtenorganisationen sehr restriktive robots.txt, die insbesondere die Suche nach Artikeln nicht zulassen. Daher beschloss ich, das Portfolio einer der großen VC-Firmen in den USA, Sequoia, zu scrapen, da deren robots.txt permissiv ist und ich außerdem denke, dass Startups und die Risikokapital-Szene im Allgemeinen sehr interessant sind.
Robots.txt
Werfen wir zunächst einen Blick auf die robots.txt von Sequoia:
Glücklicherweise erlauben sie verschiedene Zugriffsarten – mit Ausnahme von drei URLs, was für unsere Zwecke ausreichend ist. Wir werden dennoch eine Crawl-Verzögerung von 15-30 Sekunden zwischen den einzelnen Anfragen einbauen.
Als Nächstes wollen wir uns die Daten ansehen, die wir abrufen wollen. Wir interessieren uns für das Portfolio von Sequoia, also ist https://www.sequoiacap.com/companies/ die URL, die wir suchen.
Die Unternehmen sind übersichtlich in einem Raster angeordnet, so dass sie recht einfach zu scrapen sind. Wenn Sie auf die Seite klicken, werden die Details zu jedem Unternehmen angezeigt. Beachten Sie auch, wie sich die URL in Abbildung 4 ändert, wenn Sie auf ein Unternehmen klicken! Dies ist besonders für Anfragen wichtig.
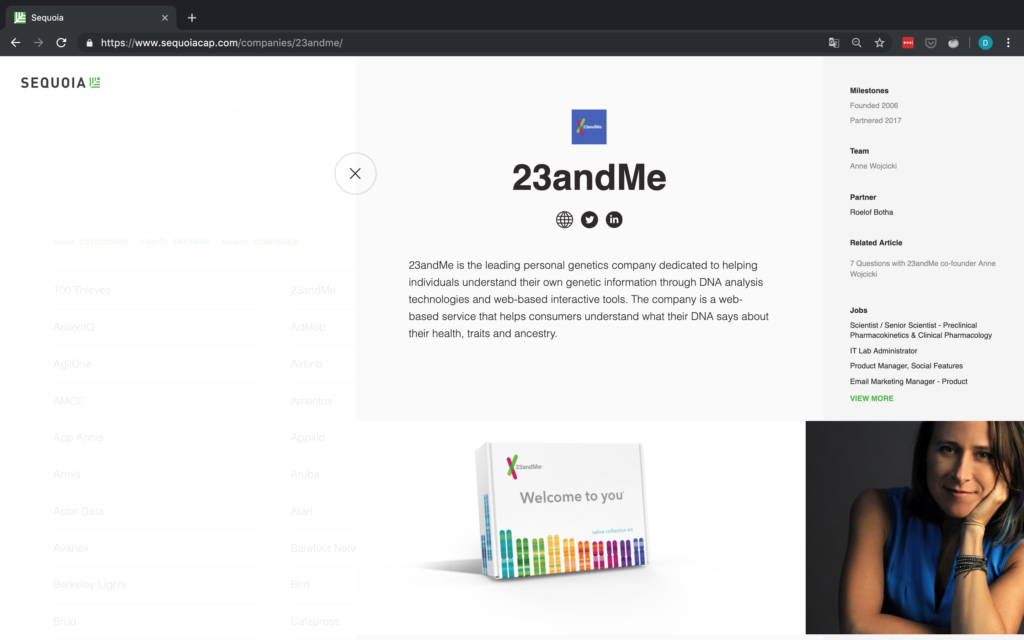
Ziel ist es, die folgenden grundlegenden Informationen über jedes Unternehmen zu sammeln und sie als CSV-Datei auszugeben:
-
Name des Unternehmens
-
URL des Unternehmens
-
Beschreibung des Unternehmens
-
Meilensteine
-
Team
-
Partner
Wenn eine dieser Informationen für ein Unternehmen nicht verfügbar ist, fügen wir stattdessen einfach die Zeichenfolge „NA“ ein.
Zeit, mit der Inspektion zu beginnen!
Scraping Prozess
Name des Unternehmens
Wenn man sich das Raster ansieht, sieht es so aus, als ob die Informationen über jedes Unternehmen in einem div-Tag mit der Klasse companies _company js-company enthalten sind. Wir sollten also in der Lage sein, mit BeautifulSoup nach dieser Kombination zu suchen.
Damit fehlen aber immer noch alle anderen Informationen, d.h. wir müssen irgendwie auf die Detailseiten der einzelnen Unternehmen zugreifen. In Abbildung 5 oben hat jedes Unternehmen ein Attribut namens „data-url“. Für 100 Thieves zum Beispiel hat onclick den Wert /companies/100-thieves/. Das ist genau das, was wir brauchen!
Jetzt müssen wir nur noch dieses data-URL-Attribut für jedes Unternehmen an die Basis-URL anhängen (die einfach https://www.sequoiacap.com/ ist), und schon können wir eine weitere Anfrage senden, um die Detailseite jedes Unternehmens aufzurufen.
Schreiben wir also etwas Code, um zunächst den Firmennamen zu ermitteln und dann eine weitere Anfrage an die Detailseite jedes Unternehmens zu senden: Ich werde hier Code schreiben, der mit Text durchsetzt ist. Ein vollständiges Skript finden Sie in meinem .
Zunächst kümmern wir uns um alle Importe und richten alle Variablen ein, die wir benötigen. Wir senden auch unsere erste Anfrage an die Basis-URL, die das Raster mit allen Unternehmen enthält und einen BeautifulSoup-Parser instanziiert.
Nachdem wir uns um die grundlegende Buchführung gekümmert und das Wörterbuch eingerichtet haben, in dem wir die Daten auslesen wollen, können wir mit dem eigentlichen Auslesen beginnen, indem wir zunächst die in Abbildung 5 gezeigte Klasse analysieren. Wie in Abbildung 5 zu sehen ist, müssen wir, nachdem wir das „div“-Tag mit der passenden Klasse ausgewählt haben, zu dessen erstem „div“-Unterelement gehen und dann dessen Text auswählen, der dann den Namen des Unternehmens enthält.
URL des Unternehmens
Für die URL sollten wir nur in der Lage sein, Elemente anhand ihrer Klasse zu finden und dann das erste Element auszuwählen, da es scheint, dass die Website immer der erste soziale Link ist. Die Inspector-Ansicht ist in Abbildung 6 zu sehen.
Aber halt – was ist, wenn es keine sozialen Links gibt oder die Website des Unternehmens nicht angegeben ist? Für den Fall, dass die Website nicht angegeben wird, wohl aber eine Social-Media-Seite, betrachten wir diesen Social-Media-Link einfach als die De-facto-Website des Unternehmens. Wenn überhaupt keine sozialen Links vorhanden sind, müssen wir einen NA anhängen. Aus diesem Grund überprüfen wir explizit die Anzahl der gefundenen Objekte, da wir nicht auf das href-Attribut eines Tags zugreifen können, das nicht existiert. Ein Beispiel für ein Unternehmen ohne URL ist in Abbildung 7 dargestellt.
p-Tag, der die Unternehmensbeschreibung enthält, keine zusätzlichen identifier. Daher sind wir gezwungen, zuerst auf den „div“-Tag darüber zuzugreifen und dann zum p -Tag mit der Beschreibung zu gehen und dessen text
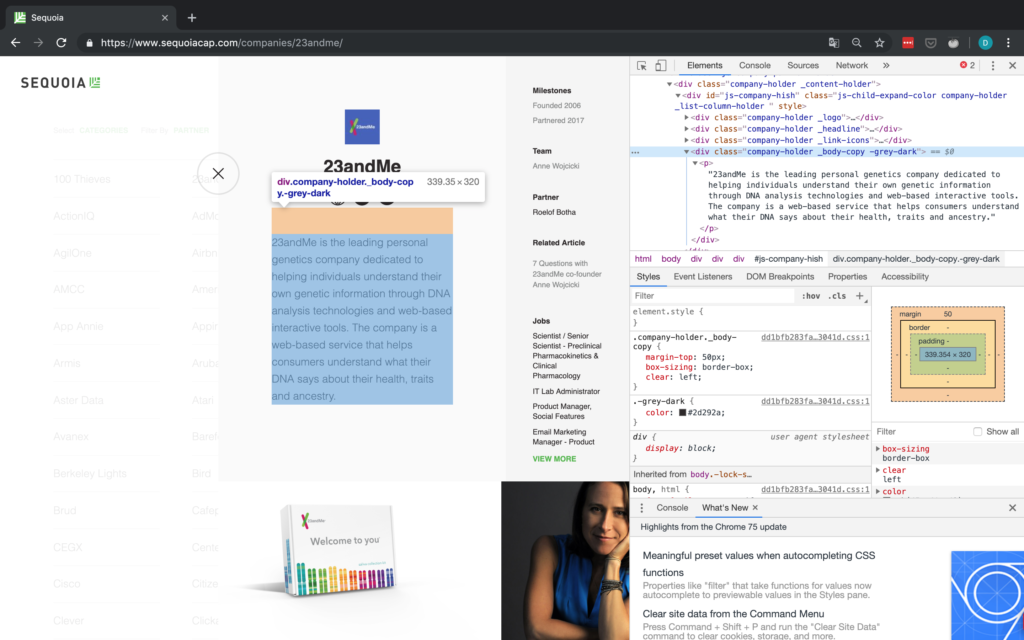
Die letzten drei Elemente befinden sich alle in derselben Struktur und können daher auf dieselbe Weise aufgerufen werden. Wir werden einfach den Text des übergeordneten Elements abgleichen und uns dann von dort aus nach unten vorarbeiten.
Da die spezifischen Textelemente keine guten Erkennungsmerkmale aufweisen, wird der Text des übergeordneten Elements abgeglichen. Sobald wir den Text haben, gehen wir zwei Ebenen nach oben, indem wir das Attribut parent verwenden. Dies bringt uns zum div-Tag, das zu dieser spezifischen Kategorie gehört (z.B. Meilensteine oder Team). Nun müssen wir nur noch zum ul-Tag hinuntergehen, das den eigentlichen Text enthält, an dem wir interessiert sind, und seinen Text abrufen.
Ein Problem bei der Verwendung der Textübereinstimmung ist die Tatsache, dass nur exakte Übereinstimmungen gefunden werden. Dies ist in Fällen von Bedeutung, in denen die gesuchte Zeichenfolge auf verschiedenen Seiten leicht unterschiedlich sein kann. Wie Sie in Abbildung 10 sehen können, gilt dies für unsere Fallstudie hier für den Text Partner. Wenn einem Unternehmen mehr als ein Sequoia-Partner zugewiesen ist, lautet die Zeichenfolge „Partners“ statt „Partner“. Daher verwenden wir bei der Suche nach dem Element „Partner“ ein REGEX, um dies zu umgehen.

Zu guter Letzt ist nicht garantiert, dass alle gewünschten Elemente (d.h. Meilensteine, Team und Partner) tatsächlich für jedes Unternehmen verfügbar sind. Bevor wir also ein Element auswählen, suchen wir zunächst alle Elemente, die mit der Zeichenfolge übereinstimmen, und überprüfen dann die Länge. Wenn es keine übereinstimmenden Elemente gibt, fügen wir NA an, andernfalls erhalten wir die erforderlichen Informationen.
Für einen Partner gibt es immer ein Element, so dass wir davon ausgehen, dass keine Partnerinformationen verfügbar sind, wenn es ein oder weniger Elemente gibt. Ich glaube, der Grund dafür, dass immer ein Element mit einem Partner übereinstimmt, ist die in Abbildung 11 gezeigte Option „Filter nach Partner“. Wie Sie sehen, ist beim Scraping oft eine Iteration erforderlich, um mögliche Probleme mit Ihrem Skript zu finden.
Auf die Festplatte schreiben
Zum Abschluss fügen wir alle Informationen zu einem Unternehmen an die Liste an, zu der es in unserem Wörterbuch gehört. Dann konvertieren wir dieses Wörterbuch in einen Pandas DataFrame, bevor wir es auf die Festplatte schreiben.
Geschafft! Wir haben soeben alle Portfolio-Unternehmen von Seqouia und Informationen über sie gesammelt.
* Nun, zumindest alle auf ihrer Website, ich glaube, sie haben ihre eigenen Websites für z.B. Indien und Israel.
Werfen wir einen Blick auf die Daten, die wir gerade gescraped haben:
Sieht gut aus! Wir haben insgesamt 506 Unternehmen erfasst, und auch die Datenqualität sieht wirklich gut aus. Das Einzige, was mir aufgefallen ist, ist, dass einige Unternehmen zwar einen Social Link haben, dieser aber nirgendwo hinführt. In Abbildung 12 sehen Sie ein Beispiel dafür bei Pixelworks. Das Problem ist, dass Pixelworks einen Social Link hat, aber dieser Social Link enthält eigentlich keine URL (das href-Ziel ist leer) und verweist einfach auf das Sequoia-Portfolio.
Ich habe dem Skript Code hinzugefügt, um die Leerzeichen durch NAs zu ersetzen, habe aber die Daten so belassen, wie sie sind, um diesen Punkt zu illustrieren.
Fazit
Mit diesem Blogbeitrag wollte ich Ihnen eine gute Einführung in das Web-Scraping im Allgemeinen und speziell in die Verwendung von Requests und BeautifulSoup geben. Jetzt können Sie es in der freien Wildbahn einsetzen, indem Sie Daten auslesen, die für Sie oder jemanden, den Sie kennen, von Nutzen sein können. Achten Sie aber immer darauf, dass Sie sowohl die robots.txt als auch die Nutzungsbedingungen der jeweiligen Seite, die Sie scrapen wollen, lesen und respektieren.
Darüber hinaus können Sie sich auf den offiziellen Websites Ressourcen und Anleitungen zu einigen der oben genannten Methoden wie Scrapy und Selenium ansehen. Sie sollten sich auch selbst herausfordern, indem Sie einige dynamischere Seiten scrapen, die Sie nicht nur mit Requests scrapen können.
Wenn Sie Fragen haben, einen Fehler gefunden haben oder einfach nur über alles, was mit Python und Scraping zu tun hat, plaudern möchten, können Sie uns gerne unter kontaktieren.